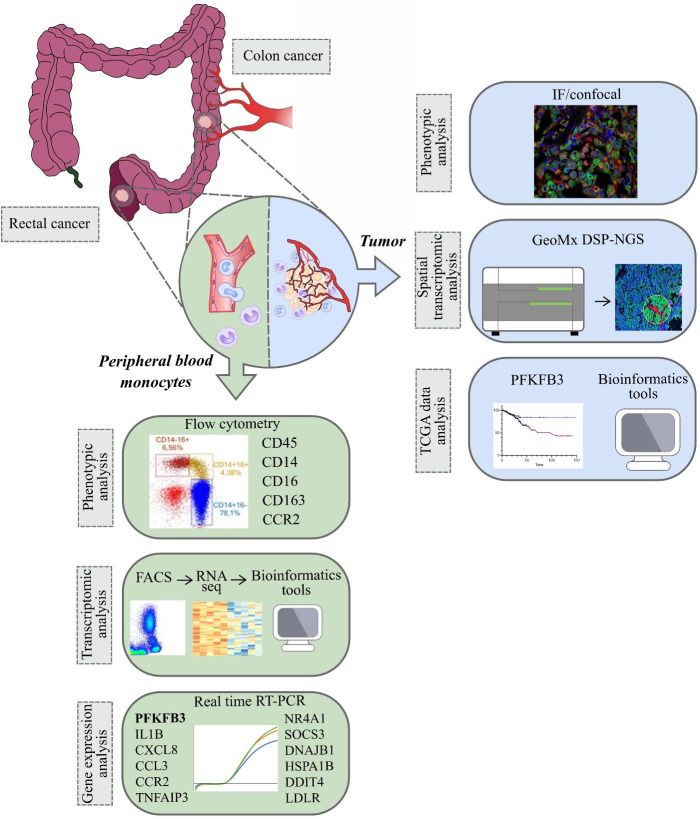
Figure 1.
The overall study design. The samples (monocytes and tumor tissue) were collected from patients with colon and rectal cancers (1). Peripheral blood monocytes were characterized by phenotypic analysis using flow cytometry, by transcriptomic analysis using RNA sequencing (Illumina technology) and by gene expression analysis using real-time RT-PCR (2). In tumors of colon and rectum, phenotypic analysis was performed with IF/confocal microscopy. Spatial transcriptomic analysis was applied using GeoMX DSP-NGS (Nanostring technology). TCGA data were obtained for survival analysis and correlations with the parameters of progression in colon and rectal cancers. All patients` cohorts and technologies are described in the methods.