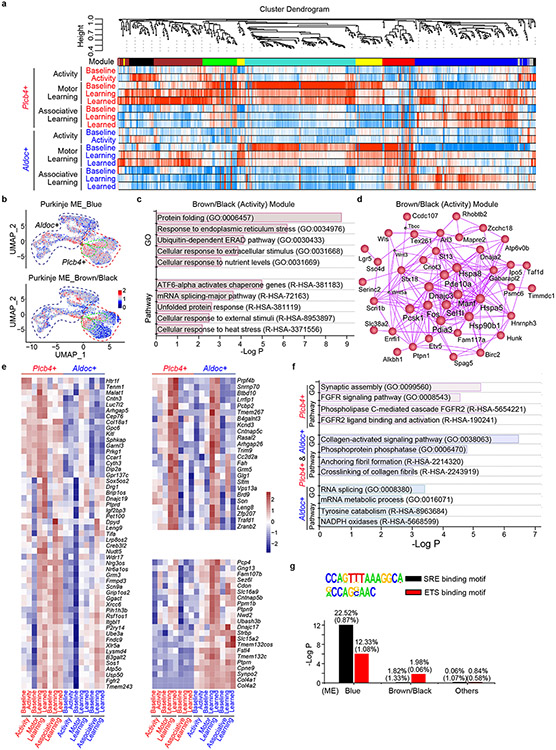
Extended Data Figure 9. Motor activity- and learning-associated gene modules in Purkinje neurons.
a, Weighted gene co-expression network analysis (WGCNA) dendrogram indicating clustering of different gene modules in Plcb4+ and Aldoc+ nuclei upon activity, motor learning or associative learning training. b, FeaturePlot showing the average expression of the activation (brown/black) and learning (blue) module genes in clusters of Plcb4+ and Aldoc+ Purkinje neurons. c, Gene ontology (GO) and Pathway analyses of the activity (brown/black) modules. d, Hub-gene network analysis of the activity (brown/black) modules. e, Enrichment of the learning genes in Plcb4+ (left panel), Plcb4+ and Aldoc+ (right up panel), or Aldoc+ (right lower panel) Purkinje neurons under motor learning and associative learning paradigms. f, GO and Pathway analyses of Plcb4+, Plcb4+ and Aldoc+, or Aldoc+ Purkinje neurons-enriched learning genes. g, Enrichment of FGFR2 signaling regulated downstream transcription factor binding motifs, serum response element (SRE) and E-Twenty Six (ETS), in the learning (blue), activity (brown/black) and other module gene sets. Enrichment of SRE and ETS in random genome DNA sequences is used as control.