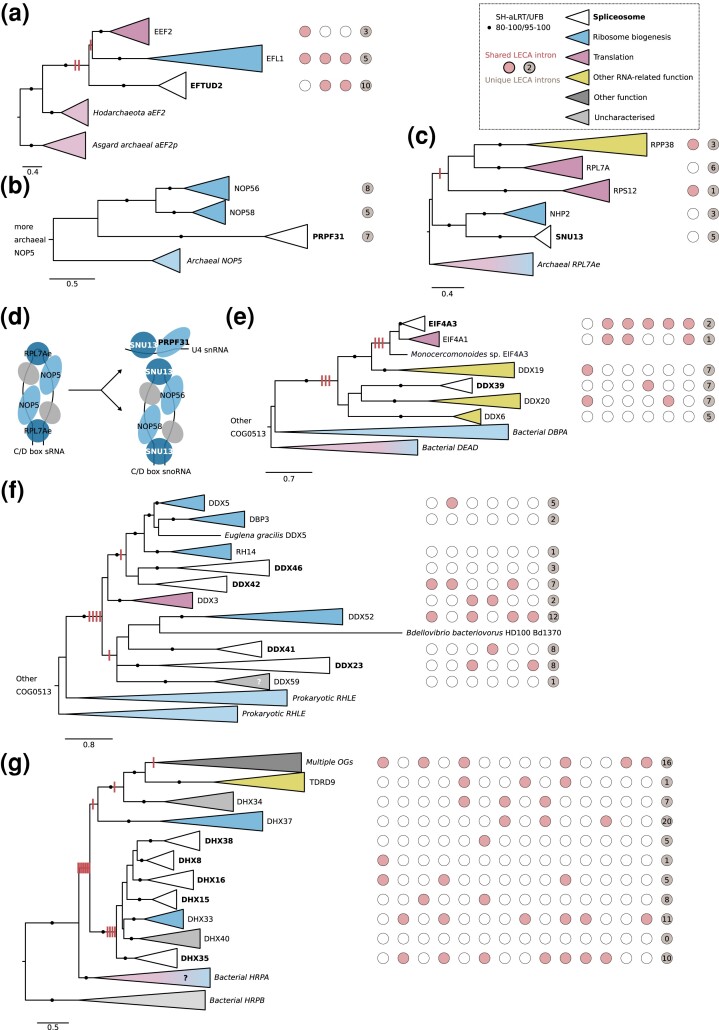
Fig. 3.
Spliceosomal proteins that originated from ribosome-related proteins. (a) Phylogenetic tree of the EF2 family. (b) Phylogenetic tree of the NOP family. (c) Phylogenetic tree of the RPL7A family. (d) Evolution of the C/D box snoRNP and U4 snRNP proteins SNU13 and PRPF31 in LECA from the C/D box sRNP in the archaeal ancestor of eukaryotes. Homologous proteins are shown in the same color. SNU13 was present in both complexes in LECA. The gray protein corresponds with fibrillarin. (e, f) Phylogenetic tree of the DDX helicase family, displaying two separate acquisitions during eukaryogenesis in two separate panels. The function of DDX59 has not been characterized, but its phylogenetic profile is similar to minor-spliceosome-specific proteins (de Wolf et al. 2021). (g) Phylogenetic tree of the DHX helicase family. (a–c, e–g) Eukaryotic LECA OGs are collapsed and colored based on their function, as are the prokaryotic clades. Introns inferred in LECA are depicted; columns with red/white circles correspond with the presence of introns at homologous positions. The gain of introns before duplications as reconstructed using Dollo parsimony is shown with red stripes on the branches. Scale bars correspond with the number of substitutions per site. Clades with significant support as assessed with the SH-like approximate likelihood ratio (SH-aLRT) and ultrafast bootstrap (UFB) values are indicated with filled circles.