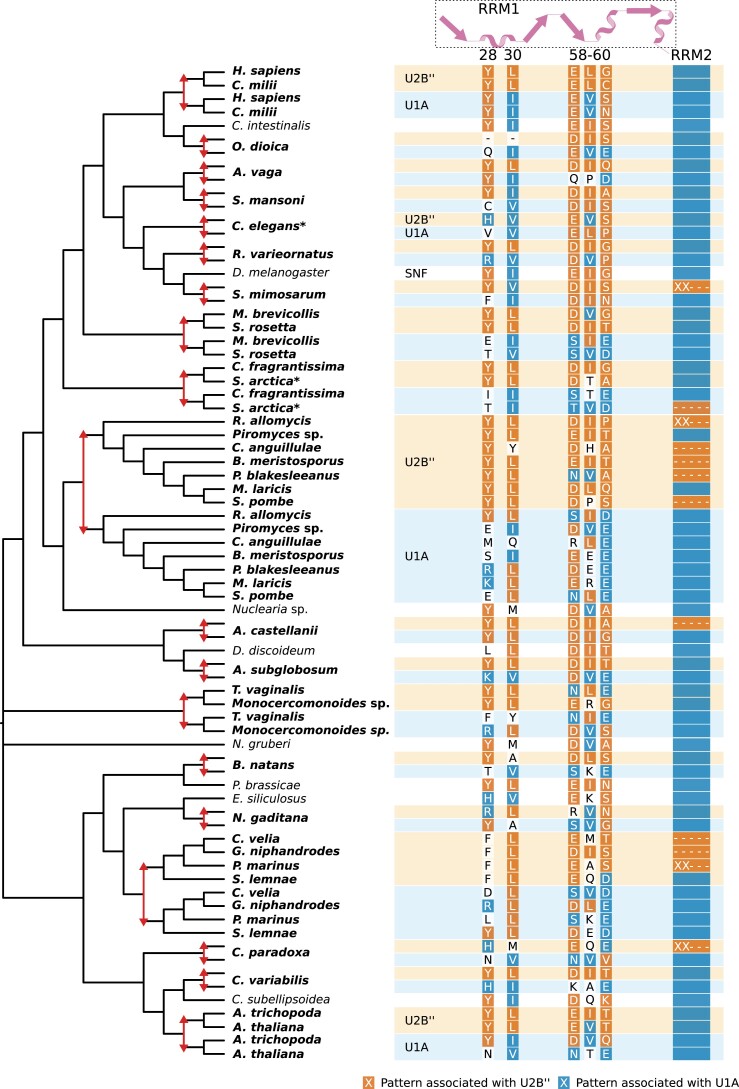
Fig. 4.
Independent gene duplications and recurrent sequence evolution in the SNF family. The reconciled tree (see supplementary fig. S6a, Supplementary Material online for the full tree) shows the positions of gene duplications (red arrows) and the species names with duplicates are in bold. The colored rectangles next to the species names correspond with the predicted fate of the duplicates. The most prominent recurrent patterns are depicted with colors corresponding with the fate this pattern is associated with. For the second RRM (RRM2), the pattern is the presence (blue bar), absence (dashes), or partial presence (XX---) of this domain. The secondary structure of the first RRM (RRM1) and the position of the patterns in the D. melanogaster sequence are shown at the top. The duplications in Sphagnum fallax and Emiliania huxleyi are not shown because the duplicates are identical for the positions that are displayed.