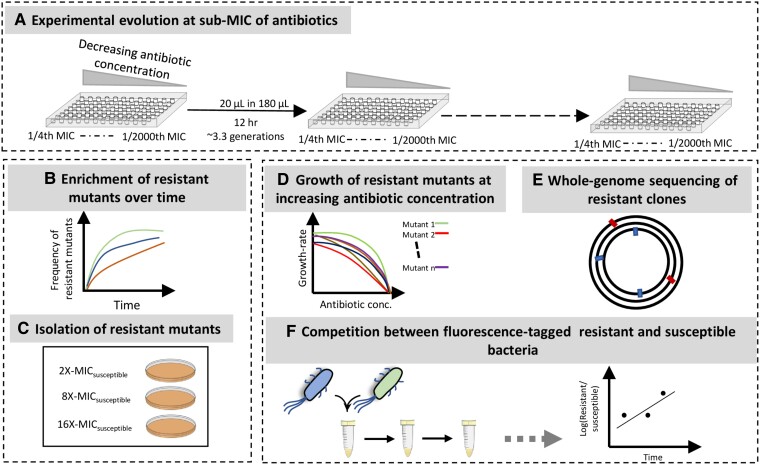
Fig. 1.
Selection of resistant bacteria at the subMICs of four antibiotics. (A) The antibiotic concentrations used in the selection experiments ranged from 1/4th to 1/2000th MICsusceptible. Eight lineages were used for each concentration of a given antibiotic, and the experiment was carried out by transferring 20 μl of overnight-grown cultures into 180 μl of fresh media every 12 h. (B, C) The frequency of resistant bacteria was measured over the course of the experiment by plating approximately 106 cells/lineage on agar plates containing antibiotic concentrations of 2 × -, 4 × -, 8 × -, and 16 × - MICsusceptible. (D–F) Resistant mutants were characterized by measuring the growth rates at different antibiotic concentrations, by identifying adaptive mutations using whole-genome sequencing, and by determining relative fitness using competition assays between fluorescently marked susceptible and resistant strains.