Fig. 5 |. Principles used to decode mechanisms of intercellular communication via expression of ligands and receptors in physically proximal cell subpopulations.
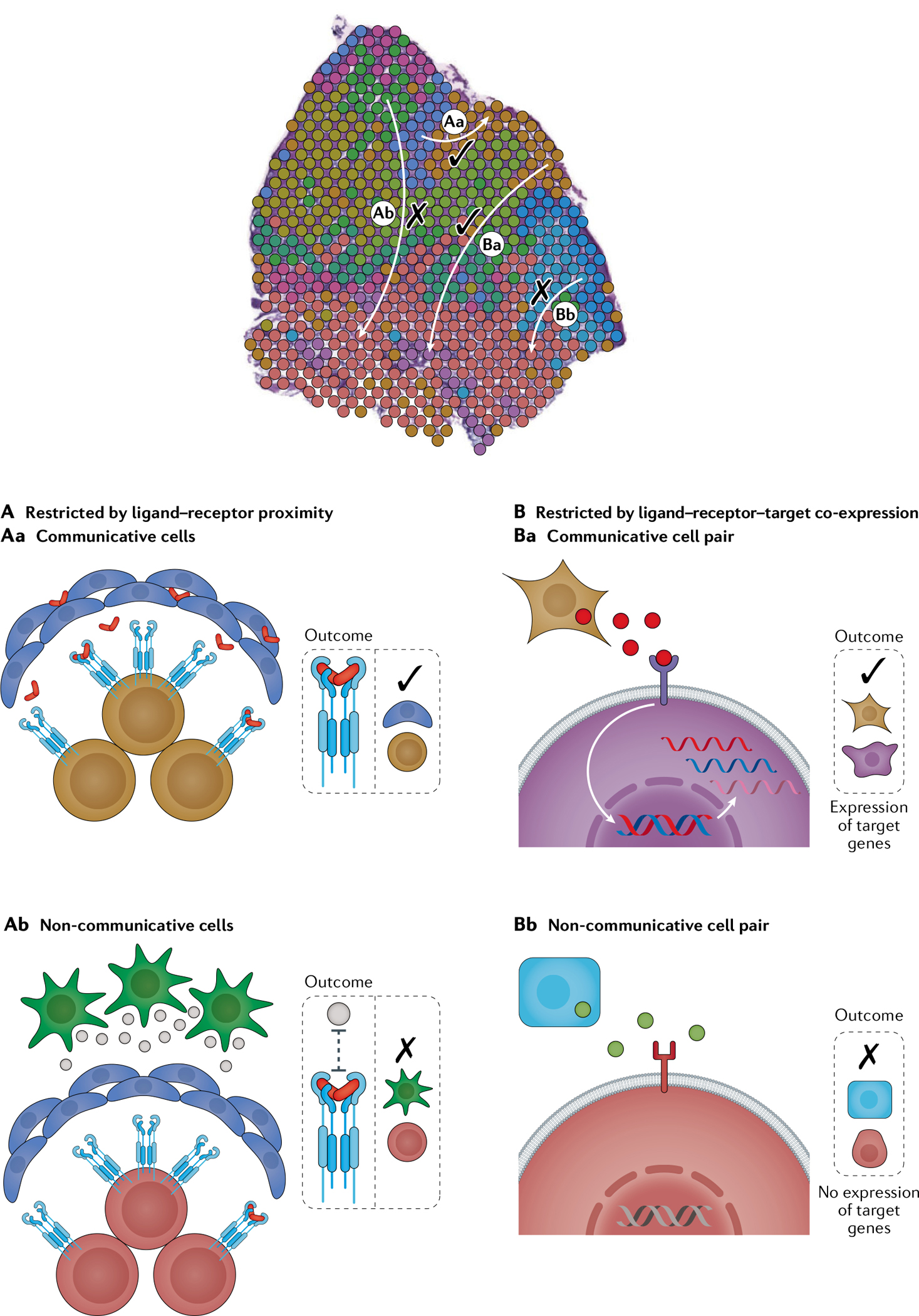
In a given tissue niche, cell subpopulations are more likely to be in communication if they are spatially proximal to each other (part A) and exhibit appropriate target gene signatures in signal-receiving cells (part B). A | Communicative cell types are established by evaluating co-localization to a given capture spot and/or expression of cell types in adjacent capture spots or cells. B | To account for longer-range communicative cell types that might be missed by the aforementioned strategies, the SpaOTsc algorithm predicts the maximum communication range for a given ligand–receptor pair through ligand–receptor target gene co-expression. Whereas ligand–receptor and ligand–receptor–target co-expression restriction can be used to establish intercellular communications from single-cell RNA sequencing (scRNA-seq) data alone, the spatial context can enhance this analysis by predicting maximum communication ranges and may disprove communications if the distance between the pairs is much farther than expected based on typical ranges recorded in the literature. Tissue cross-section adapted from REF.33, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
