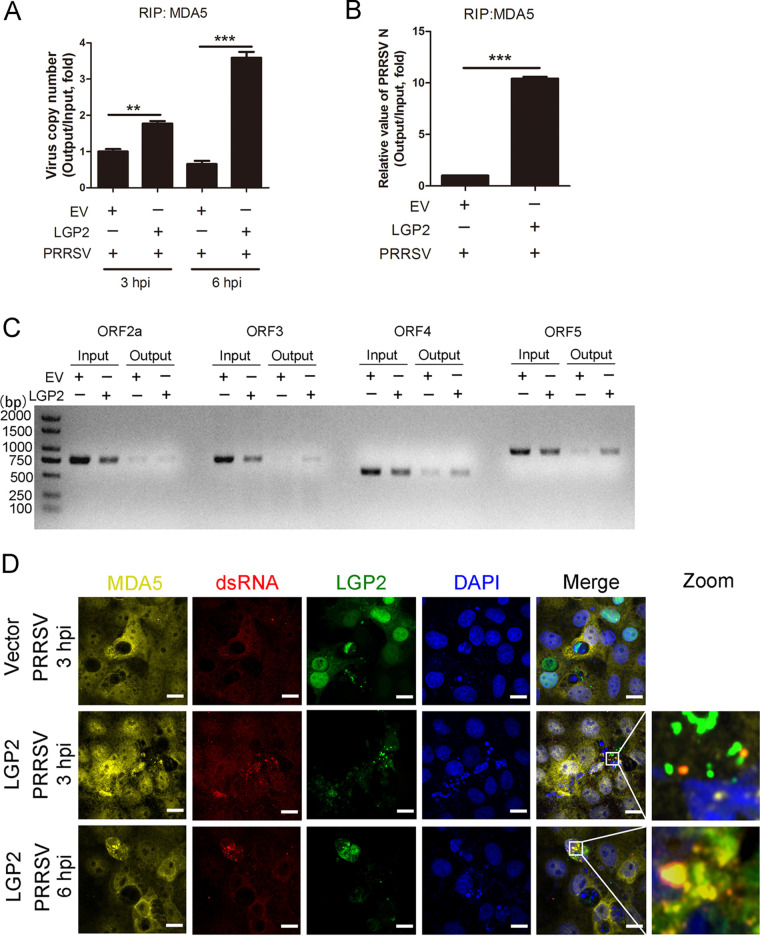
FIG 4.
LGP2 facilitated viral RNA binding to MDA5. (A) Marc-145 cells were mock transfected and transfected with LGP2 plasmids for 24 h and then infected with PRRSV (MOI = 5) for another 3 or 6 h. Cell lysates were immunoprecipitated with an MDA5 antibody, and immunoprecipitated RNA was extracted and reverse transcribed. The PRRSV copy number was detected using TaqMan real-time qPCR. The columns represent the relative value of precipitated viral RNA (Output)/total viral RNA (Input). (B) Marc-145 cells were mock transfected and transfected with LGP2 plasmids for 24 h and then infected with PRRSV (MOI = 5) for another 6 h. Cell lysates were immunoprecipitated with an MDA5 antibody, and the relative value of PRRSV N (ORF7) was detected using SYBR green qPCR. (C) The agarose gel electrophoresis of PRRSV ORF2a, ORF3, ORF4, and ORF5 PCR products after RIP assays and amplified with the corresponding primers. (D) Marc-145 cells were mock transfected and transfected with GFP-tagged LGP2 plasmids for 24 h and then infected with PRRSV (MOI = 5) for another 3 or 6 h. Immunofluorescence analysis of the colocalization among MDA5 (yellow), LGP2 (green), dsRNA (red), and DAPI (blue) is performed. Scale bar, 10 μm. The data are the results of three independent experiments (means ± the SE). Significant differences are denoted by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).