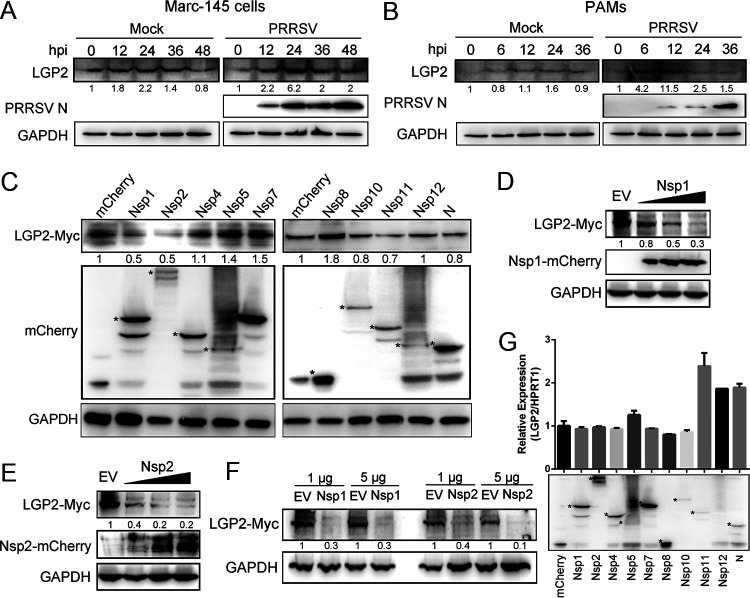
FIG 6.
PRRSV Nsp1 and Nsp2 reduce the expression of LGP2 protein. (A) Marc-145 cells were mock infected or infected with PRRSV (MOI = 1) for 0, 12, 24, 36, and 48 h. Western blot analyzed the expression of LGP2 and PRRSV N. (B) PAMs were mock infected or infected with PRRSV (MOI = 1) for 0, 6, 12, 24, and 36 h. The protein levels of LGP2 and PRRSV N were determined using Western blot analysis. (C) HEK293T cells were cotransfected with mCherry-tagged PRRSV proteins (1.5 μg) plasmids and Myc-tagged LGP2 (1.5 μg) for 24 h. Cells were collected, and the expression of LGP2-myc and mCherry-fusion viral proteins were determined using Western blot analysis. Asterisks mark the expressed mCherry-fusion proteins of PRRSV nonstructural proteins and N protein. GAPDH is shown as an internal control. (D) HEK293T cells were cotransfected with the plasmids expressing Myc-tagged LGP2 (1.5 μg) and increased doses of mCherry-tagged PRRSV Nsp1 (1, 3, and 5 μg). After 24 h of transfection, cells were lysed and subjected to Western blot analysis with anti-mCherry, anti-Myc, and anti-GAPDH antibodies. (E) HEK293T cells were cotransfected with Myc-tagged LGP2 (1.5 μg) plasmid and increased doses of mCherry-tagged PRRSV Nsp2 (1, 3, and 5 μg) plasmids for 24 h. The expression of LGP2, Nsp2, and GAPDH was determined using anti-Myc, anti-mCherry, and anti-GAPDH antibodies. (F) Marc-145 cells were cotransfected with the plasmids expressing Myc-tagged LGP2 (1.5 μg) and different doses of mCherry-tagged PRRSV Nsp1 and Nsp2 (1 and 5 μg). Western blotting was used to analyze the protein level of LGP2 and GAPDH. (G) Empty plasmid and plasmids expressing PRRSV Nsp1, Nsp2, Nsp4, Nsp5, Nsp7, Nsp8, Nsp10, Nsp11, Nsp12, and N were transfected into 3D4/21 cells, respectively. The transcript levels of LGP2 were determined using qRT-PCR. Western blotting was used to detect the expression of viral proteins. Asterisks mark the expressed mCherry-fusion proteins of PRRSV proteins. For the Western blot, the relative band density was normalized to the loading control GAPDH and then compared to the corresponding control. The data are the results of three independent experiments (means ± the SE). Significant differences are denoted by asterisks (*, P < 0.05; **, P < 0.01; ***, P < 0.001).