Figure 5.
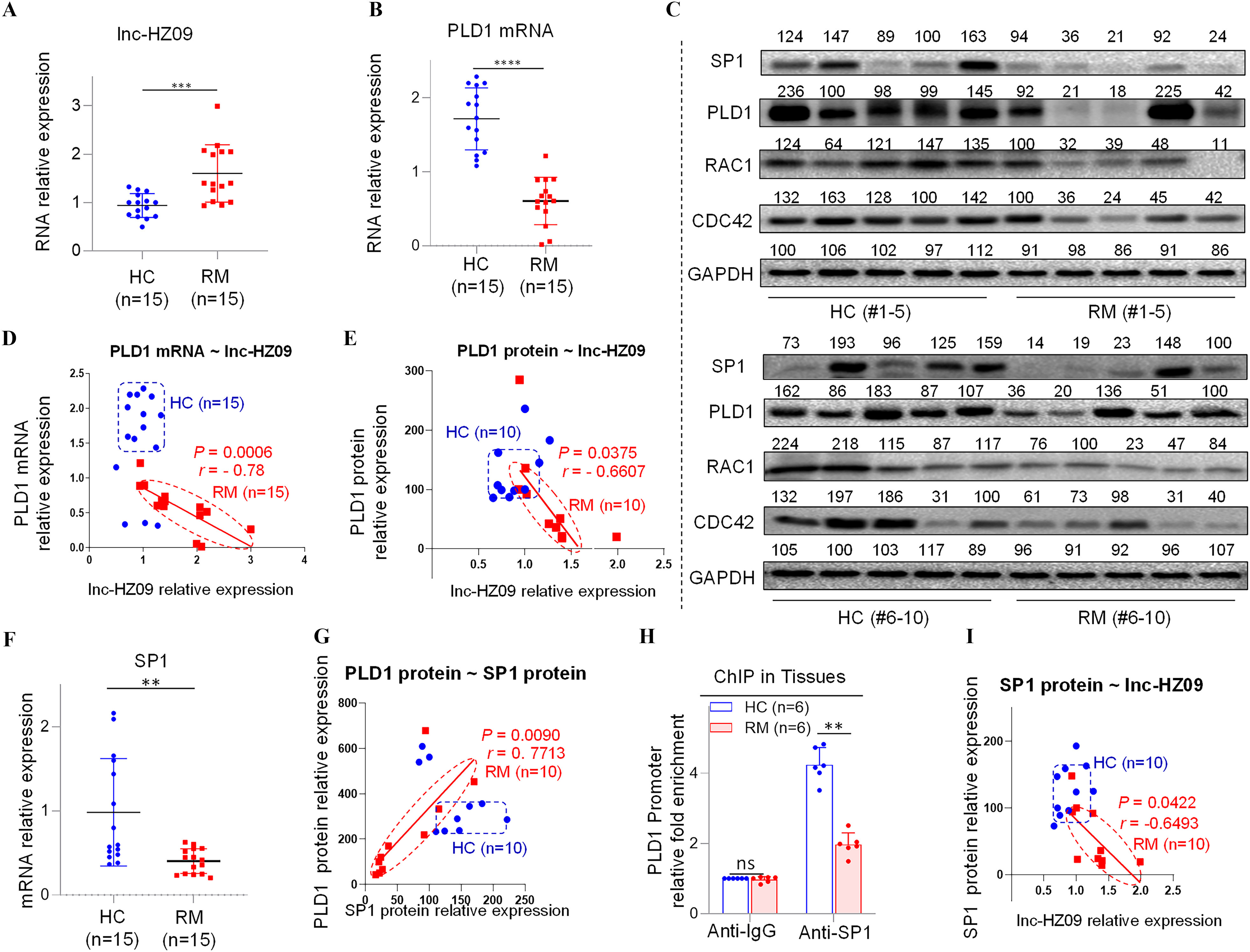
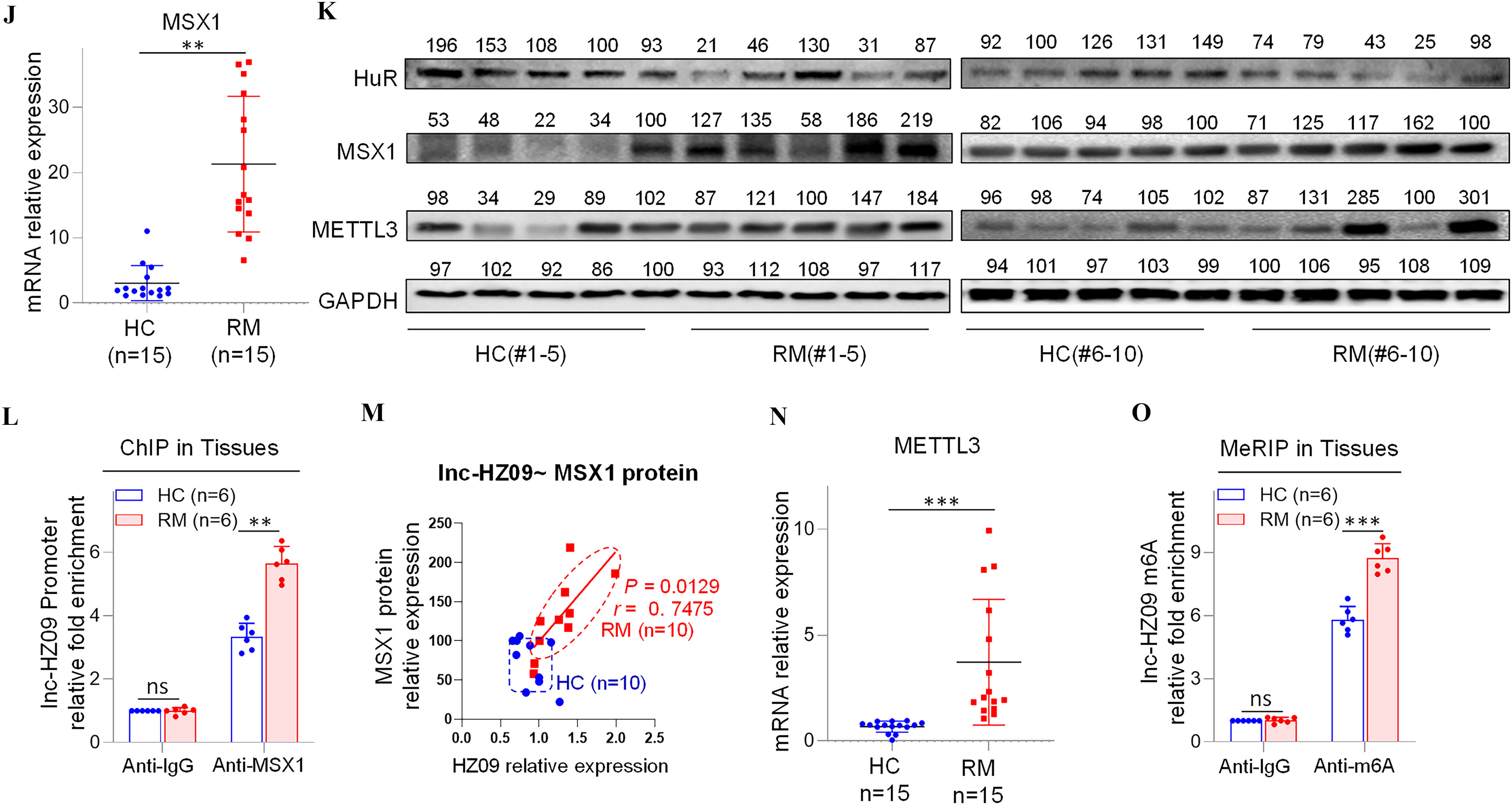
Regulation roles of lnc-HZ09 in human villous tissues. (A–B) RT-qPCR analysis of the levels of lnc-HZ09 (A) or PLD1 mRNA (B) in HC (healthy control, round) and RM (recurrent miscarriage, square) tissues (each ). (C) Western blot analysis of the protein levels of SP1, PLD1, RAC1, and CDC42 in HC and RM tissues (each ), with GAPDH as internal standard. The relative intensity of each band was quantified, and their levels were shown in Figure S10C. (D) The correlation between PLD1 mRNA levels and lnc-HZ09 levels in HC (round) and RM (square) tissues (each ). (E) The correlation between PLD1 protein levels and lnc-HZ09 levels in HC (round) and RM (square) groups (each ). (F) RT-qPCR analysis (each ) of the mRNA levels of SP1 in HC (round) and RM (square) tissues (each ). (G) The correlation between the protein levels of PLD1 and SP1 in HC (round) and RM (square) groups (each ). (H) SP1 ChIP assay analysis of the relative enrichment of SP1 in the promoter region of PLD1 gene in HC and RM tissues (each ). (I) The correlation between SP1 protein levels and lnc-HZ09 levels in HC (round) and RM (square) groups (each ). (J) RT-qPCR analysis of the mRNA levels of MSX1 in HC and RM tissues (each ). (K) Western blot analysis of the protein levels of HuR, MSX1, and METTL3 in HC and RM tissues (each ), with GAPDH as internal standard. The relative intensity of each band was quantified, and their levels were shown in Figure S10H,J,K. (L) MSX1 ChIP assay analysis of the relative enrichment of MSX1 in the promoter region of lnc-HZ09 in HC and RM tissues (each ). (M) The correlation between the levels of lnc-HZ09 and the protein levels of MSX1 in HC (round) and RM (square) groups (each ). (N) RT-qPCR analysis (each ) of the mRNA levels of METTL3 in HC and RM tissues (each ). (O) MeRIP assay analysis (each ) of the levels of m6A RNA methylation on lnc-HZ09 in HC and RM tissues (each ). The summary data of these bar charts and scatter plots were shown in Excel Table S1. The DNA or RNA level in IgG group was set as “1” in all of ChIP and MeRIP assays and the middle intensity value was set to “100” in all of western blot assays. (C–E, G, I, K, M) shows representative data from three independent experiments. Data in (H, L, O) show of six independent experiments. Two-tailed Student’s -test for (A–B,F,H,J,L,N–O); Pearson analysis for (D,E,G,I,M). *, **, and ***. Note: ChIP, chromatin immunoprecipitation; GADPH; glyceraldehyde-3-phosphate dehydrogenase; HC, healthy control group; IgG, immunoglobulin G; MeRIP, methylated RNA immunoprecipitation; n, the number of biologically independent samples; ns, nonsignificance; PLD1, phospholipase D hydrolyze 1; RM, recurrent miscarriage group.
