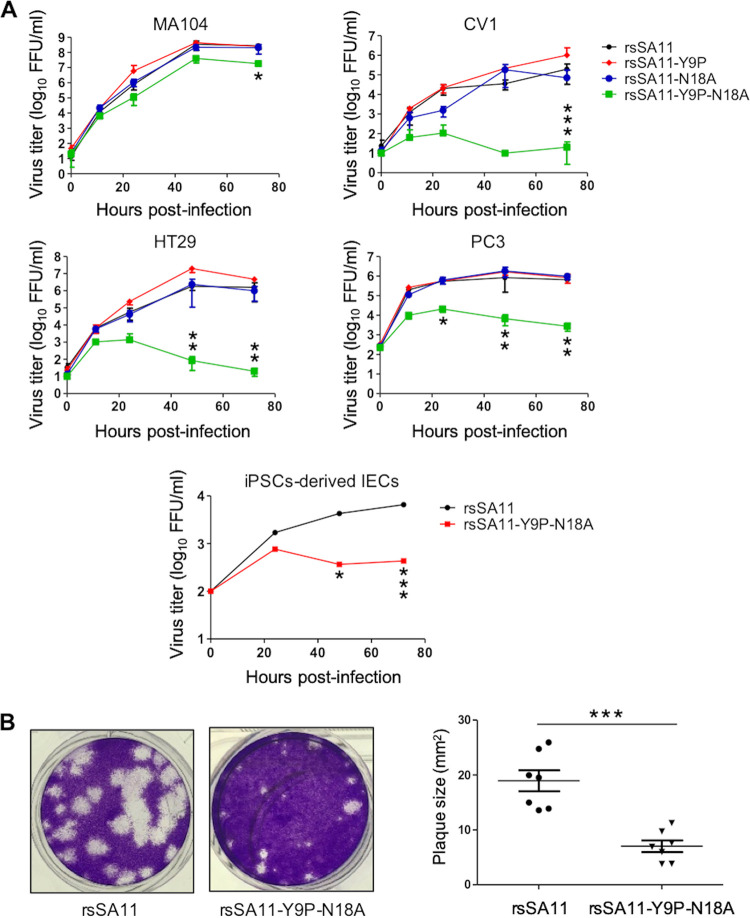
FIG 2.
Effect of NSP4 N-glycosylation on viral replication. (A) Replication kinetics of NSP4 N-glycosylation-defective recombinant viruses in multiple cell lines. MA104, CV1, HT29, and PC3 cells were infected with rsSA11, rsSA11-Y9P, rsSA11-N18A, or rsSA11-Y9P-N18A at an MOI of 0.01. Cells were collected at 0, 12, 24, 48, and 72 hpi and freeze-thawed three times at −80°C. IECs derived from human iPSCs were infected with rsSA11 or rsSA11-Y9P-N18A at an MOI of 0.005. Cells were collected at 0, 24, 48, and 72 hpi and freeze-thawed three times at −80°C. The virus titer was determined in a focus assay using MA104 cells. Foci were detected using antiserum against NSP4. Significant differences in replication between rsSA11 and rsSA11-Y9P-N18A were determined by two-way analysis of variance (ANOVA). *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Confluent MA104 cells were infected with the indicated viruses and subjected to a plaque assay. Plaque size (n = 7) was quantified in ImageJ by comparing the pixel size of the plaques with the dish size. The data are expressed as means ± the SD. The significance of the differences in plaque size between rsSA11 and rsSA11-Y9P-N18A was determined using a Mann-Whitney U test. ***, P < 0.001.