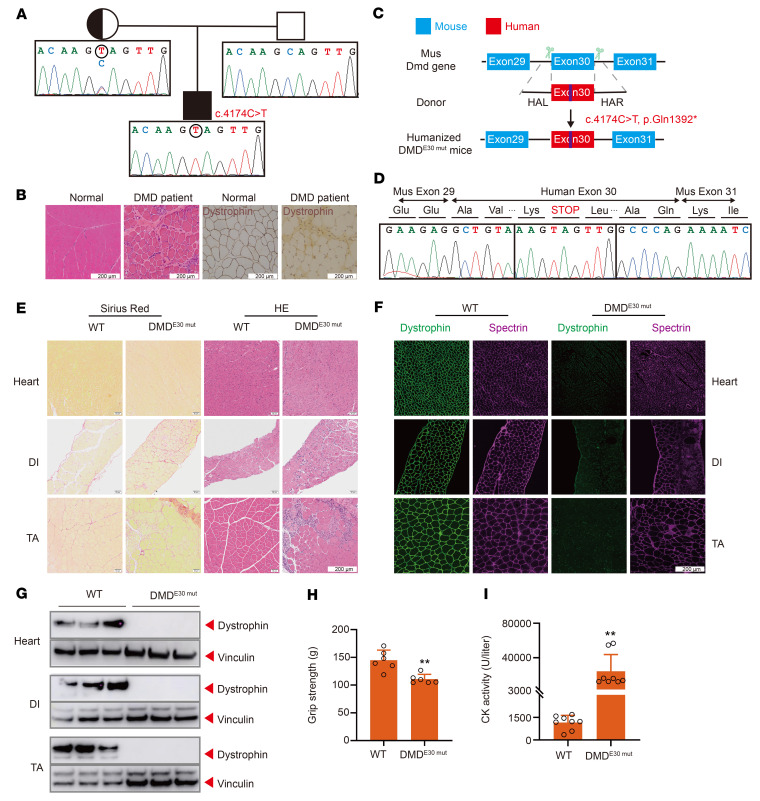
Figure 1. Establishment and characterization of a humanized DMD mouse model.
(A) Pedigree of patient with DMD (proband) with a nonsense mutation p.Gln1392*. Squares represent males; circles represent females. (B) Histological analysis of left biceps muscle from normal and proband. Dystrophin (MilliporeSigma, D8168) is shown in brown. (C) Strategy for generating humanized DMD mouse model. CRISPR/Cas9 editing using 2 sgRNAs flanking an exon was used to delete mouse Dmd exon 30 and replace it with human DMD exon 30 carrying the nonsense mutation. (D) RT-PCR products from muscle of DMDE30mut mice were sequenced to validate the exon 30 mutation. (E) Sirius red staining and H&E staining of TA, DI, and heart muscle of WT and DMDE30mut mice. (F) Dystrophin immunohistochemistry from indicated muscles of WT and DMDE30mut mice. Dystrophin (Abcam, ab15277) and spectrin (Millipore, MAB1622) are shown in green and magenta, respectively. (G) Western blot confirming the absence of dystrophin in indicated muscle tissues. (H) WT and DMDE30mut mice were subjected to forelimb grip strength testing to measure muscle performance (n = 6). (I) Serum CK, a marker of muscle damage and membrane leakage, was measured in WT and DMDE30mut mice (n = 8). All mice were 8 weeks old at the time of the experiment. Data are represented as mean ± SEM. Each dot represents an individual mouse. **P < 0.01 using unpaired 2-tailed Student’s t test. Scale bars: 200 μm.