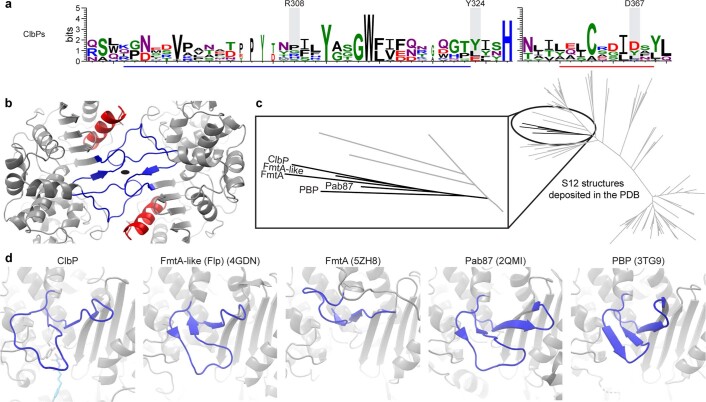
Extended Data Fig. 9. ClbP dimerizes through loops that are not highly conserved.
a, Sequence logo representing conservation of dimer interface regions highlighted in panel b on the structure and in Supplementary Figure 3 on the sequence among 15 ClbP homologs from colibactin biosynthetic clusters. Residues predicted to be important for dimerization are not strongly conserved, suggesting that the mode of dimerization we observe in E. coli ClbP may be an adaptation of Proteobacterial ClbP. b, ClbP dimer interface highlighting the α8-β11 loop region (residues 296-324; blue) and α11 helix (357-372; red). c, Unrooted sequence similarity tree of S12 homologs with structures deposited in the PDB. The inset details the homologs in the same clade as ClbP. d, Equivalent views of the α8-β11 loop region (blue) in the structures of homologs in the same clade as ClbP. The α8-β11 loop region are highly variable in structure in the S12 homologs. These regions only mediate formation of a dimer in ClbP and FmtA (PDB ID: 5ZH8), but the dimer geometry is different and only the ClbP dimer has the active sites of the two subunits facing each other on either side of the substrate-binding cavity. The catalytic triad and product analog of ClbP are shown as sticks for context.