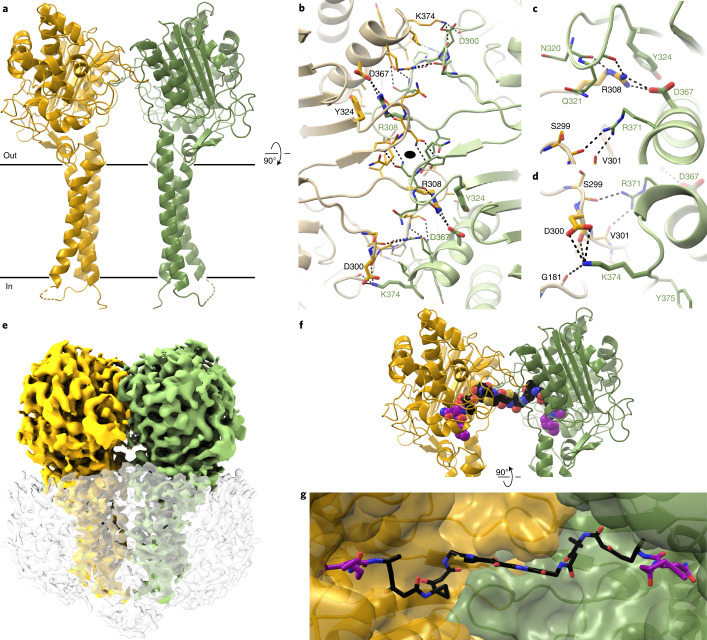
Fig. 4. ClbP forms a dimer that accommodates pseudodimeric precolibactin.
a, ClbP dimer observed in the crystal-packing interactions from a plane perpendicular to the cell membrane, denoted as black lines. b, Orthogonal view of the dimer interface looking from the periplasm to the inside of the cell. The interface forms around a two-fold crystallographic symmetry axis (black oval) and consists of a pair of interlocking loops that contribute both hydrophobic and polar interactions. The largest predicted energetic contributors to stabilizing this interface are interactions formed among residues R308, Y324 and D367 (shown as thick sticks). All other residues participating in the interface are shown as thin sticks. c,d, Detailed view of interactions mediated by R308 (c) and K374 (d). e, 3D reconstruction obtained from cryo-EM analysis of wild-type ClbP. Density colored to correspond to each subunit, and the detergent micelle is shown as a transparent surface with dust hidden for clarity. f,g, Model of precolibactin binding to the ClbP dimer obtained by individually docking fragments of the molecule (Supplementary Fig. 5). Precolibactin can straddle both subunits of the ClbP dimer such that the prodrug motifs at both ends can each bind a different active site simultaneously. Views of precolibactin binding to the dimer as seen from a plane perpendicular to the membrane (f) as well as to the surface of the cavity subtended by the dimer (g). Note that the docked molecule contains hexanoyl chains in place of the natural tetradecanoyl (or ‘C14’) chains of the myristoyl groups.