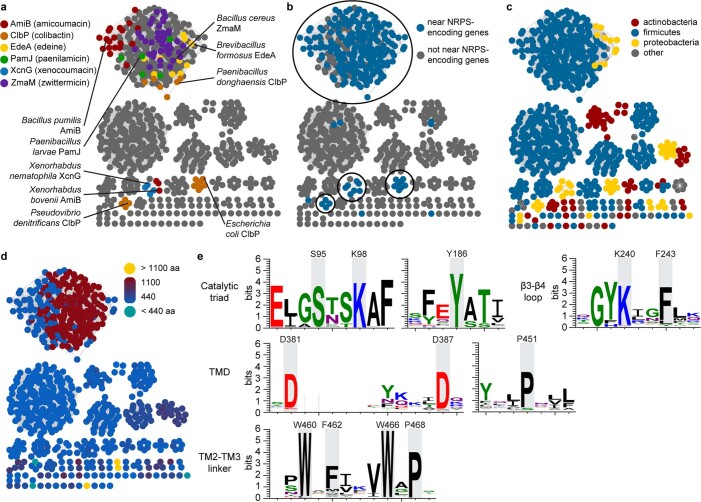
Extended Data Fig. 1. Identification and sequence conservation of prodrug-activating homologs of ClbP.
a, Sequence similarity network (SSN) for 730 ClbP homologs, colored by identified BGC (if any). Peptidases involved in amicoumacin, edeine, paenilamicin, and zwittermicin biosynthesis cluster together, along with the newly identified probable Gram-positive colibactin producers. The Gammaproteobacterial ClbPs are split into two distinct subsets, one comprising close relatives of the sequences found in Pseudovibrio strains56–59 and the other representing ClbPs from BGCs with canonical architecture in Escherichia species (with homologs from Erwinia60, Frischella, Gilliamella, and Serratia strains56, among others). Similarly, AmiB homologs in Xenorhabdus strains cluster with XcnG sequences rather than Gram-positive AmiBs. Intriguingly, the genomes of some strains – such as the edeine-producing B. formosus NF2 – appear to have multiple biosynthetic gene clusters containing authentic prodrug peptidases with potentially distinct activities. b, SSN colored to highlight proximity (within a ±10 gene neighborhood) of the peptidase gene to a gene containing both NRPS A and C domains, as a proxy for the presence of a NRPS module playing a ClbN-like role. The only SSN clusters in which this condition was met were clusters containing homologs of known prodrug peptidases (circled in black). All our sequence conservation analyses were performed using these clusters. c, SSN colored by phylum, highlighting that prodrug-activating peptidases are most common among Firmicutes, with some spread into Proteobacteria (as seen with the ami, clb, and xcn BGCs) and into Actinobacteria. d, The prodrug-activating peptidase SSN is colored by amino acid sequence length to emphasize that a large subset of sequences (including EdeA, PamJ, and ZmaM homologs) are much longer. This can be attributed to fusion with a second domain with homology to components of an ABC transporter, commonly annotated as a cyclic peptide transporter. However, Gram-positive AmiB and ClbP sequences (and other unidentified but related proteins) lack this additional domain and more closely resemble E. coli ClbP, X. bovenii AmiB, and XcnG. e, Sequence logos built from the alignment of 271 candidate prodrug-activating peptidases detail sequence conservation of the catalytic triad and of periplasmic-TMD interface and intra-TMD positions discussed in the main text.