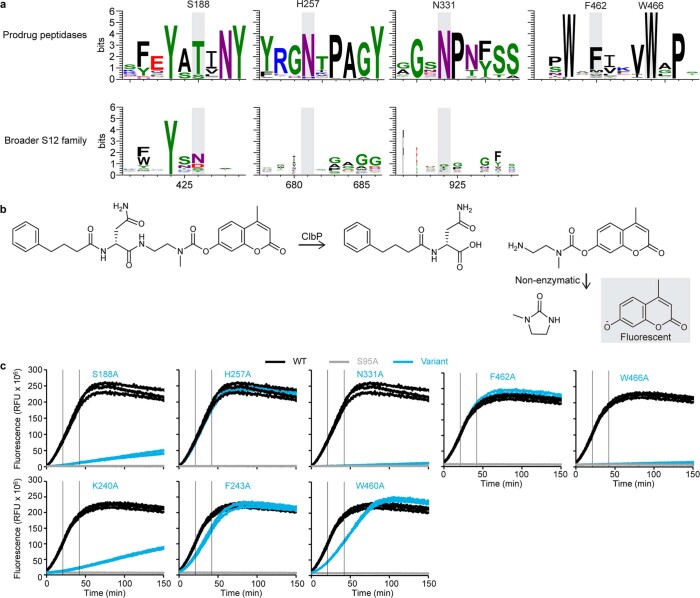
Extended Data Fig. 3. Activity of ClbP variants with mutations to conserved substrate-binding residues measured using a fluorogenic assay.
a, Sequence logos representing conservation of N-acyl-d-asparagine binding residues among 271 aligned sequences of prodrug-activating homologs (top), compared to logos built from an alignment of 901 representative sequences from the broader S12 family downloaded from the MEROPS database. b, Fluorogenic activity assay used to measure the peptidase activity of ClbP variants20. Cleavage of the synthetic substrate probe by ClbP generates an intermediate which then undergoes a non-enzymatic cyclization reaction to yield the active fluorophore (gray box). c, Curves of the raw fluorescence versus time for different ClbP variants with point mutations at residues of interest that interact with the substrate (top row) or form notable interdomain (K240A and F243A) or intra-TMD (W460) interactions (see Extended Data Fig. 1e for the corresponding sequence logos). Each panel represents triplicate measurements for the indicated variant (cyan). For comparison, the corresponding triplicate measurements for wild-type ClbP (black) and catalytically inactive S95A (gray), measured in the same experiment, are reproduced on each graph. The two gray vertical lines bound the data used for calculating the normalized hydrolysis rates in Fig. 2.