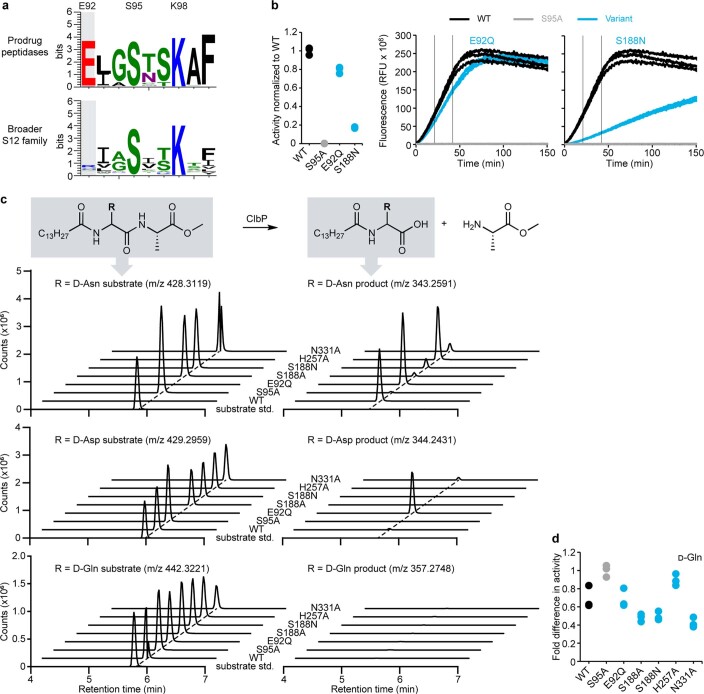
Extended Data Fig. 4. Wild-type ClbP and variants with mutations at d-asparagine binding residues cannot process substrate with an N-acyl-d-glutamine prodrug motif.
a, Sequence logo representing conservation of E92, which stabilizes the orientation of d-asparagine specificity residue N331. b, Normalized hydrolysis rates calculated from activity assays performed with d-asparagine binding mutants E92Q and S188N (left). Triplicate fluorescence activity measurements for each mutant (cyan) are shown with triplicates for wild-type ClbP (black) and catalytically inactive S95A (gray) collected in the same experiment (right). c, Assay measuring activity of mutants for dipeptide substrates containing d-asparagine, d-aspartate, or d-glutamine prodrug motifs by LC–MS detection of cleaved product. Extracted Ion Chromatogram (EIC) traces of the [M + H]+ ion for each of the substrates tested (left) and the expected ClbP cleavage product (right). d, None of the d-asparagine binding mutants process substantial amounts of the d-glutamine substrate, as indicated by the lack of a difference in activity between the catalytically deficient S95A and any of the other variants (n = 3 independent experiments).