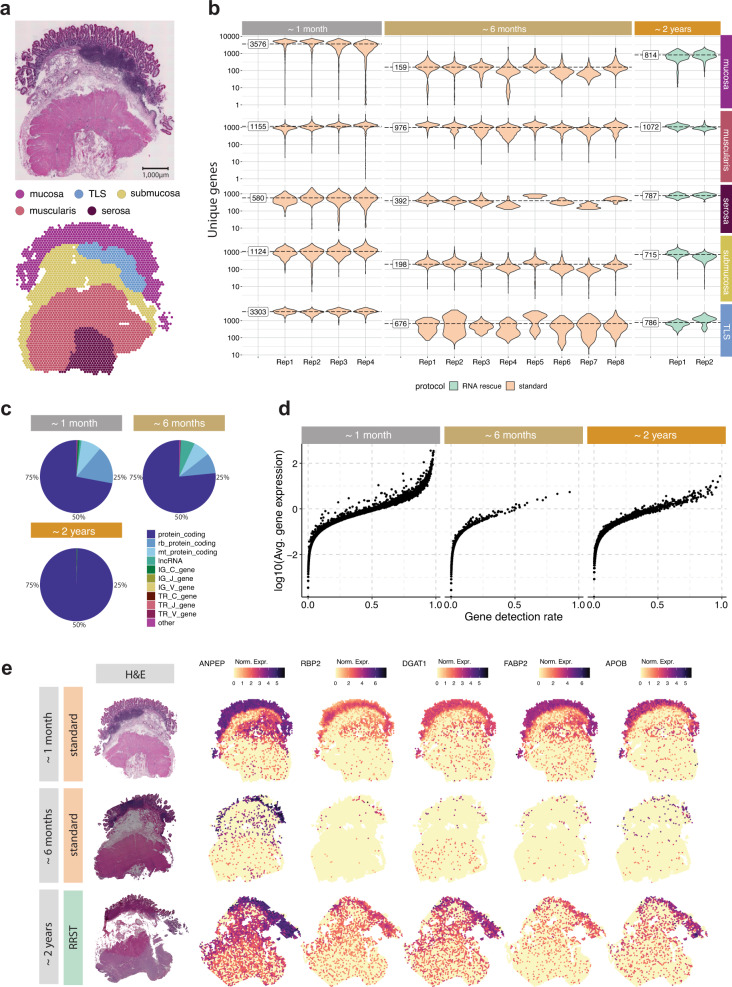
Fig. 4. Comparison between RRST and standard Visium on an adult human small intestine sample over time.
a Representative H&E image (top) and spots colored by five major tissue regions (bottom): mucosa, TLS, submucosa, muscularis, and serosa. TLS, Tertiary Lymphoid Tissue. The full small intestine dataset consisted of 14 tissue sections collected from the same specimen at different time points. Only sections collected at the last point were processed with RRST, while the other sections were processed with standard Visium protocol. b Overview of data quality in the five annotated tissue regions over time, visualized by violin plots of the number of unique genes per spot. The time points represent the approximate storage time after sample collection: ~1 month, ~6 months, and ~2 years. Replicates obtained for each time point are shown on the x-axis. The fill color of the violin plots indicates the applied protocol. For each time point, labels on the left side of the violin plots represent the average over all replicates. c RNA biotype content for the three datasets visualized as a pie chart. Proportions represent the UMI counts detected for each biotype. The targeted RRST data include protein coding, immunoglobulin, and T-cell receptor transcripts. d Mean-detection rate relationship in the mucosa for data collected at the three different time points. The y-axis shows log10-transformed average number of UMIs for each gene, and the x-axis shows the detection rate for each gene. The detection rate is defined as the fraction of spots where the gene is detected. e Spatial visualization of five enterocyte markers. Each row represents one selected tissue section from each time point with their corresponding H&E image in the leftmost column. Spot colors represent normalized gene expression.