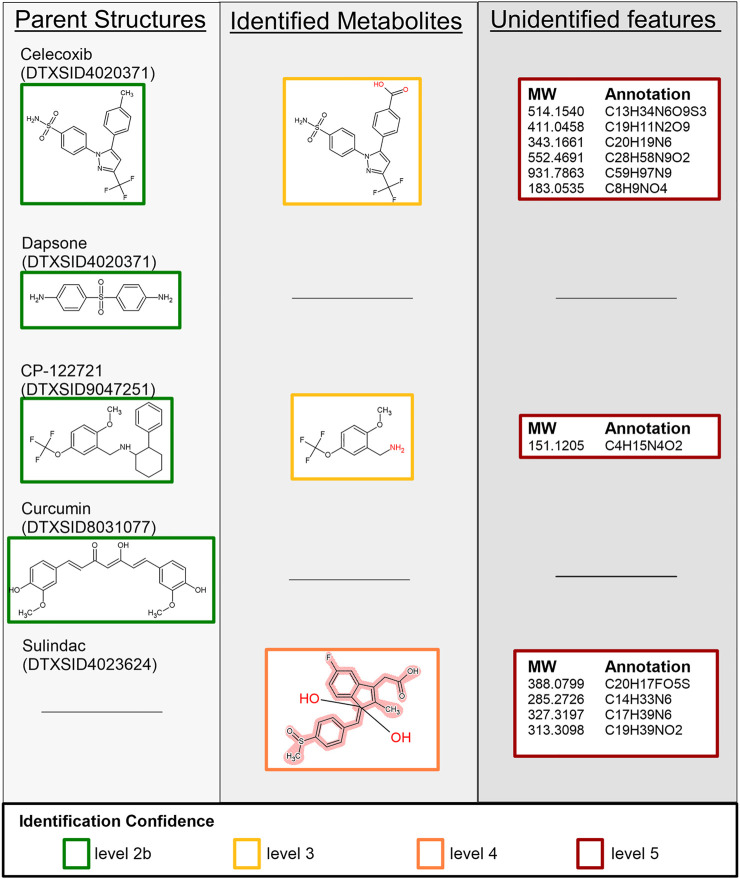
FIGURE 6.
Summary of identified and unidentified features extracted from the MS1 and MS2 data for celecoxib, dapsone, CP-122721, curcumin, and sulindac. Boxes surrounding individual structures denote the level of confidence in that assignment as described in Section 2.5.4, and the black line indicates that no features were identified to correspond to the respective row and column. Because SSA is not optimized for any one compound, our workflow relied on occurrence of chemical signals in replicate samples and time-correlated changes in signal to filter the signals. In some cases, real chemical detections may have been lost. For example, no features were identified that corresponded to the sulindac parent structure but multiple isomeric sulindac metabolites are represented with a Markush structure. Unidentified features include the molecular weight (MW) and formula annotation.