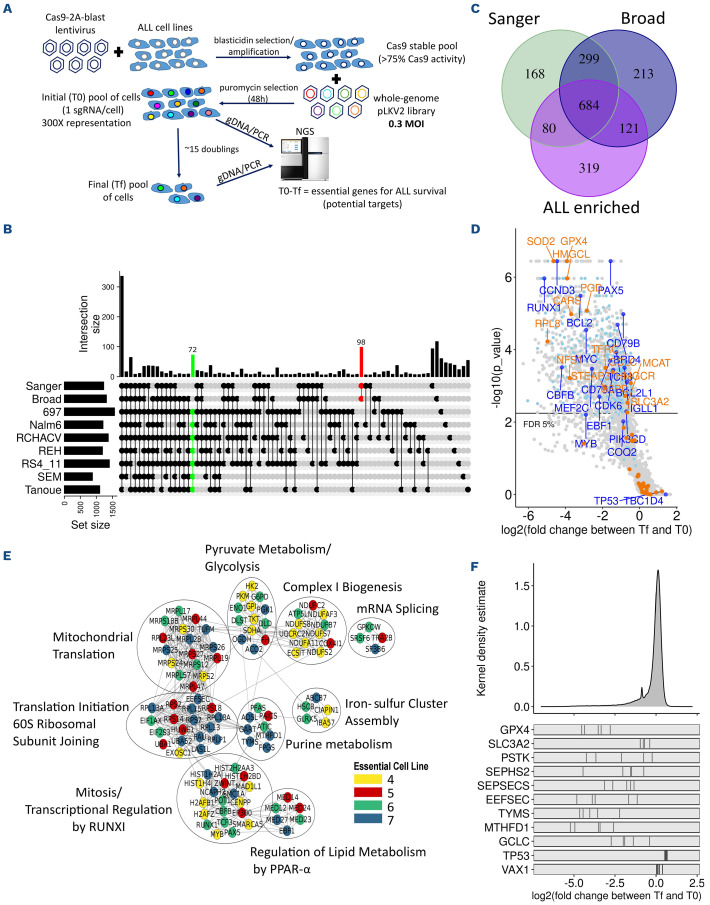
Figure 1.
Whole-genome CRISPR/Cas9 screens of acute lymphoblastic leukemia (ALL) cell lines identify ALL essential genes. (A) Schema of B-ALL screening pipeline. (B) Significant genes set intersection of 7 B-ALL cell line CRISPR screen results with UpSet, “Broad” and “Sanger” essential gene list at false discovery rate (FDR) <0.05. The intersections with more than 5 genes are shown. (C) Overlap between B-ALL essential (>4/7 cell lines, FDR <0.05) genes, “Broad” and “Sanger” core essential genes. (D) Representative Volcano plot of 697 cells CRISPR screen results with FDR <0.05. B-ALL-known dependencies and B-ALL-enriched essential genes are highlighted in dark blue and in light blue, respectively. Genes regulating ferroptosis induction are highlighted in orange) Gene Network Analysis of B-ALL-enriched essential genes where nodes represent genes and edges represent proteinprotein interactions between genes. Node color represents the number of ALL cell lines essential for the given gene. (F) Log2 fold change distribution of single-giude RNA (sgRNA) in the 7 ALL screened cell lines. sgRNA log2 fold change for ferroptosis related genes are illustrated at bottom of the panel. VAX1, negative control. TP53 was enriched in several B-ALL cell lines. Tf: final time points; M.O.I: multiplicity of infection.