Figure EV3. Comparison of DNA binding and unwinding by WRN and Sgs1 DNA helicases.
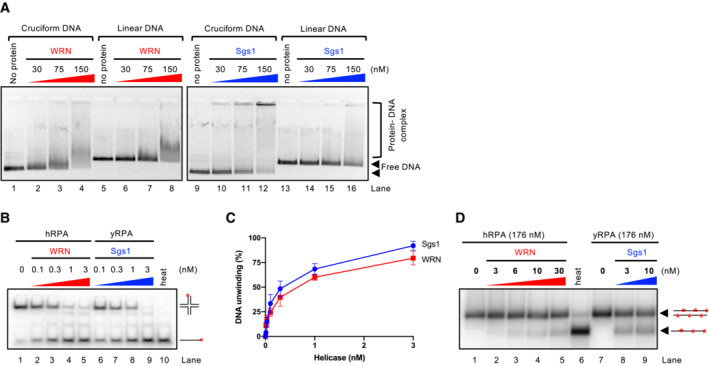
- Representative electrophoretic mobility shift assay with WRN and Sgs1, using either pUC19 harboring the random inverted repeat‐based cruciform structure (circular), or linearized pUC19. The samples were run on a 0.8% unstained native agarose gel. The gel was stained after the run with GelRed.
- Representative helicase assay with increasing concentrations of WRN and Sgs1, using an oligonucleotide‐based Holliday junction as a substrate. Reactions were supplemented with either human or yeast RPA (15 nM) and analyzed by 10% native acrylamide gel electrophoresis. Red asterisk indicates the position of the radioactive label.
- Quantitation of DNA unwinding from assays such as in (B). Averages shown; n ≥ 3; error bars, SEM.
- Representative helicase assay with increasing concentrations of WRN and Sgs1, using a 2.2 kbp‐long dsDNA substrate. Reactions were supplemented with either human or yeast RPA and analyzed by electrophoresis on a 1% agarose gel.
