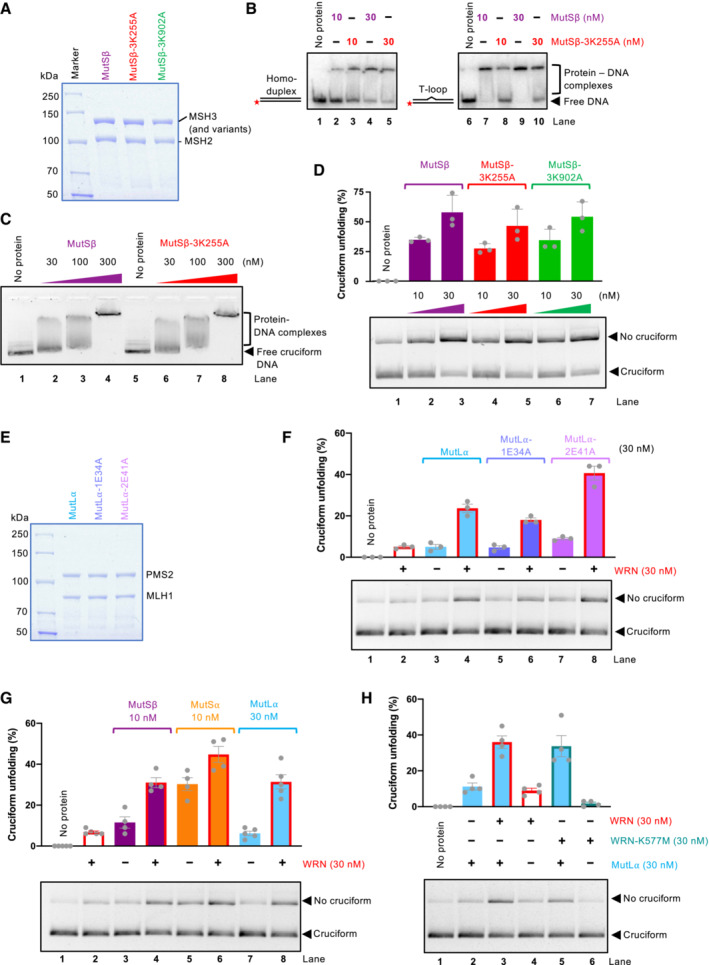
Representative polyacrylamide gel showing recombinant MutSβ, MutSβ‐3K255A (MSH3 with mutation K255A, deficient in mismatch recognition), and MutSβ‐3K902A (MSH3 with mutation K902A, ATPase‐deficient). The gel was stained with Coomassie Brilliant blue.
Electrophoretic mobility shift assay with MutSβ, MutSβ‐3K255A, and MutSβ‐3K902A, using either dsDNA (50 bp) or dsDNA bearing one extrahelical T, as a substrate. 6% native acrylamide gel was used.
Electrophoretic mobility shift assays with MutSβ and MutSβ‐3K255A, using circular pUC19 with the AT repeat cruciform structure, as a substrate. 0.8% native agarose gel was used.
Cruciform unfolding assays with MutSβ, MutSβ‐3K255A, and MutSβ‐3K902A. Bottom, representative experiments; top, quantitation, averages shown; n = 3 technical replicates; error bars, SEM.
Representative polyacrylamide gel showing recombinant MutLα, MutL⍺‐1E34A (MLH1 with mutation E34A, ATPase‐deficient), and MutL⍺‐2E41A (PMS2 with mutation E41A, ATPase‐deficient). The gel was stained with Coomassie Brilliant blue.
Unfolding of TA cruciform by WRN, MutLα, and its ATPase‐deficient variants. Bottom, representative experiments; top, quantitation; averages shown; n = 3 technical replicates; error bars, SEM.
Unfolding of TA cruciform by WRN and MMR proteins. Bottom, representative experiments; top, quantitation; averages shown; n = 4 technical replicates; error bars, SEM.
Unfolding of TA cruciform by MutLα and wild‐type WRN or helicase dead WRN‐K577M. Bottom, representative experiments; top, quantitation, averages shown; n = 4 technical replicates; error bars, SEM.