Figure 5. zRbm14 functions by enriching maternal RNA into its phase‐separated condensates.
-
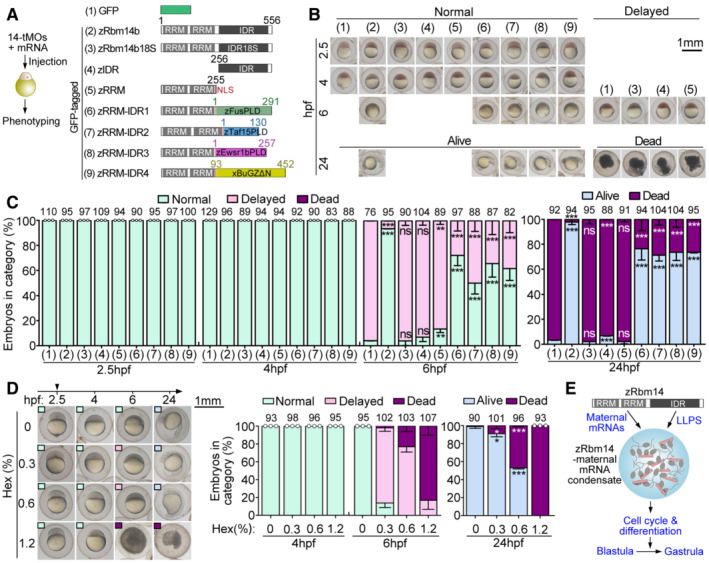
AExperimental scheme and diagrams of constructs used in the rescue experiments. A nuclear localization signal (NLS) was added to the zRRM and zRRM‐IDR1‐4 constructs to maintain their nuclear localization capacity. Prion‐like domains (PLD) of zFus, zTaf15, and zEwsr1b (Wang et al, 2018) and the low complexity region of Xenopus BuGZ (xBuGZΔN; Jiang et al, 2015) were used to replace the IDR of zRbm14b (zIDR; Xiao et al, 2019). RRM, RNA‐recognition motif.
-
B, CPhenotypes of embryos. Typical embryos, cropped from representative experimental groups in Fig EV2G, are shown in (B). Their phenotypes served as criteria for quantifications in (C). Only GFP‐positive embryos viable at 2.5 hpf were collected and imaged for analyses. Total embryo numbers analyzed are listed over histograms.
-
DHexanediol (Hex) treatment from 2.5 hpf blocked the blastula development. Representative images are presented for each group of embryos, with phenotypes indicated by insets of colored boxes. Total embryo numbers analyzed are listed over histograms.
-
EA summarizing model. zRbm14 binds to maternal mRNAs through the N‐terminal RRM‐containing region and phase separates into cytoplasmic condensates through its IDR. Such maternal mRNA‐zRbm14 condensates facilitate proper cell cycle and differentiation for embryonic development into the gastrula stage.
Data information: Quantification results (mean ± SD) were from three independent experiments. Sample dots are included for results with no error bars and thus excluding t‐test. Unpaired two‐sided Student's t‐test against control group (group 1): ns, no significance; **P < 0.01; ***P < 0.001.
Source data are available online for this figure.
