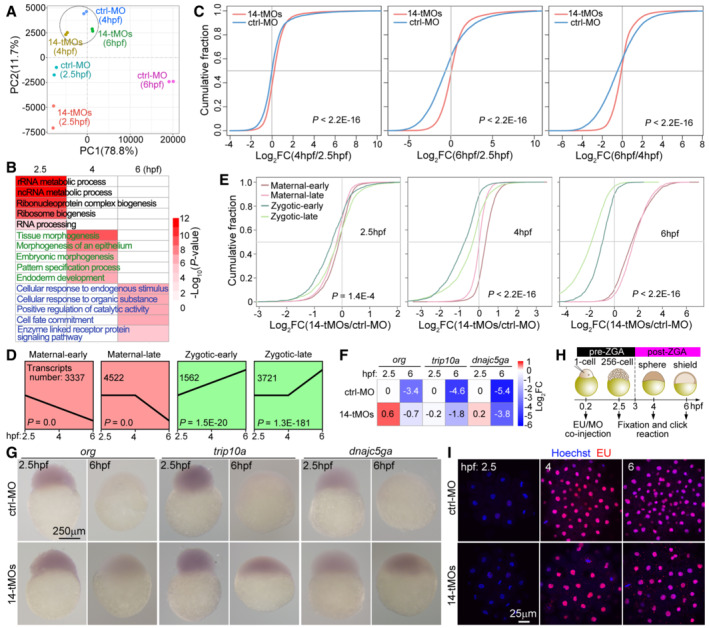
Figure 6. Depletion of maternal zRbm14 hinders maternal RNA clearance and MZT.
-
AVariance in gene expression profiles of control (ctrl‐MO) and maternal zRbm14 (14‐tMOs) morphants. Principal component analysis (PCA) was performed using two biological replicates.
-
BRepresentative gene ontology (GO) biological processes enriched for differential transcripts between control and maternal zRbm14 morphants.
-
CCumulative distribution showing fold changes (FCs) in transcript abundance in control morphants (blue curves) and maternal zRbm14 morphants (red curves) between the indicated stages. Transcript abundances were averaged from two independent experiments. P values were calculated using two‐sided Wilcoxon and Mann–Whitney tests.
-
DExpression patterns of four clusters of representative transcripts, categorized from RNA‐seq results of ctrl‐MO embryos. The transcripts in the maternal‐early and the zygotic‐early clusters respectively displayed continual degradation or accumulation from 2.5 hpf, whereas those in the maternal‐late and zygotic‐late clusters respectively started degradation or accumulation from 4 hpf.
-
ECumulative distribution showing FCs of transcript abundance of the indicated clusters between control and maternal zRbm14 morphants.
-
FRelative transcript levels of three typical maternal genes. FCs were calculated relatively to the levels in 2.5‐hpf ctrl‐MO embryos. org, oogenesis‐related; trip10a, thyroid hormone receptor interactor 10a; dnajc5ga, DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a.
-
GWhole‐mount in situ RNA hybridization of org, trip10a, and dnajc5ga. A representative embryo is shown for each group of at least 26 embryos.
-
H, IMaternal zRbm14 morphants displayed normal ZGA onset. MO(s) and 5‐ethynyl uridine (EU) were co‐injected and treated as illustrated (H). The diagrams are not drawn to scale. EU incorporated into nascent transcripts were visualized through a click reaction. Hoechst 33342 was used to counterstain the nucleus. A representative maximum intensity‐projected confocal micrograph, taken from the animal pole, is shown for each group of 10 embryos (I).
Data information: P values of the sequencing data in (C) and (E) were calculated by using two‐sided Wilcoxon and Mann–Whitney tests. P values in (D) were calculated by using Short Time‐series Expression Miner (STEM) software. A P value of < 0.05 is considered statistically significant.
Source data are available online for this figure.
