Fig. 4. Biased agonists that induce Gαi:β-arrestin complex formation induce similar conformations of β-arrestin.
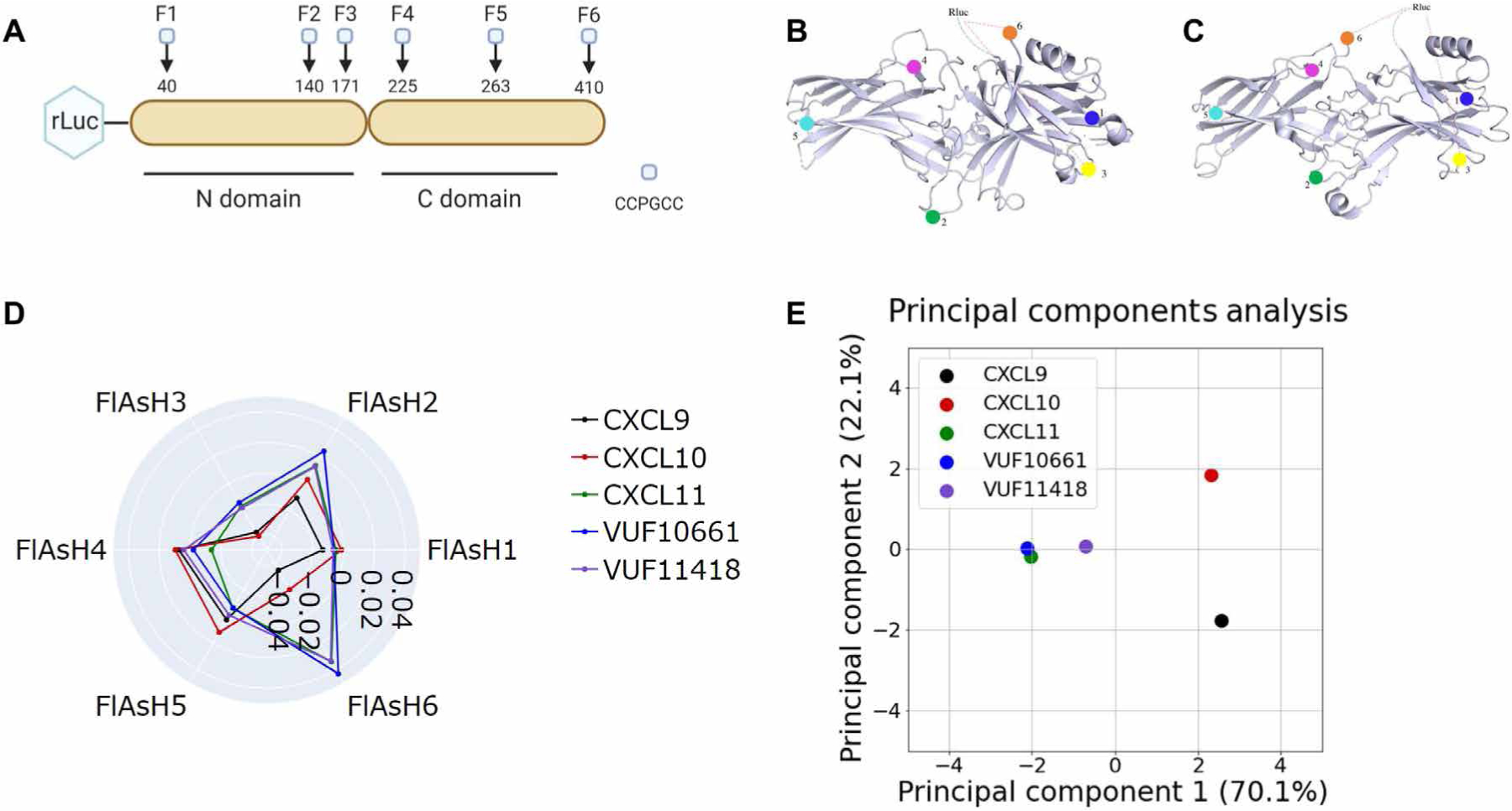
(A) The RLuc–β-arrestin2–FlAsH1 to RLuc–β-arrestin2–FlAsH6 reporters (F1 to F6) have the amino acid motif CCPGCC inserted after residues 40, 140, 171, 225, 263, and 410 of β-arrestin2. This motif acts as a dipole acceptor to assess conformational changes in β-arrestin2. (B and C) The positions of the FlAsH binding motifs are highlighted in the inactive (B) and active (C) structures of β-arrestin1. (D) HEK293N cells were transfected with plasmids encoding CXCR3 and one of the RLuc–β-arrestin2–FlAsH reporters (FlAsH1 to FlAsH6). The cells were then treated with 100 nM CXCL9, 100 nM CXCL10, 100 nM CXCL11, 1 μM VUF10661, 1 μM VUF11418, or vehicle. The radar plot depicts the intramolecular net BRET ratio calculated from subtracting the vehicle from treatment groups. (E) PCA was used to assess biased agonist–induced β-arrestin conformation similarities; computed principal components are visualized with the top two principal components. Principal component 1 contributes to 70% of the observed variation, and principal component 2 contributes to 22% of the observed variation. Points denote the composite response of a single ligand at varying doses. Data are means of three to five independent experiments.
