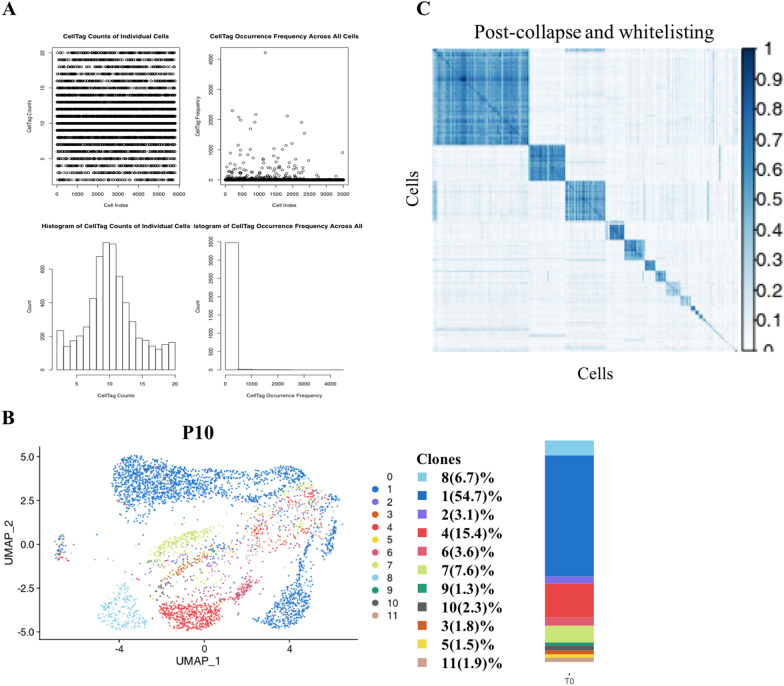
Fig. 3.
(A) Generation of the CellTag whitelist. Following single cell RNA sequencing, CellTags were first extracted from the raw sequencing files and the number of CellTags that entered each cell were calculated. Cells were not uniquely labeled with high confidence if there was only one CellTag per cell. Hence, we let each cell be tagged with 2–20 CellTags to increase the specificity of each cell. Through detection, we found that most of the labeled cells expressed 5–15 CellTags. (B) Visual clone distribution of the P10 CellTag data: projection of CellTag clones onto the UMAP plot. The histogram shows the proportion of cells from different clones. (C) CellTag signatures were extracted for each cell, and the overlap of CellTag signatures between cells were assessed using the Jaccard similarity analysis. Hierarchical clustering of cells based on each cell’s Jaccard similarity index with other cells to define ‘block’ of cells that originated from a single clone