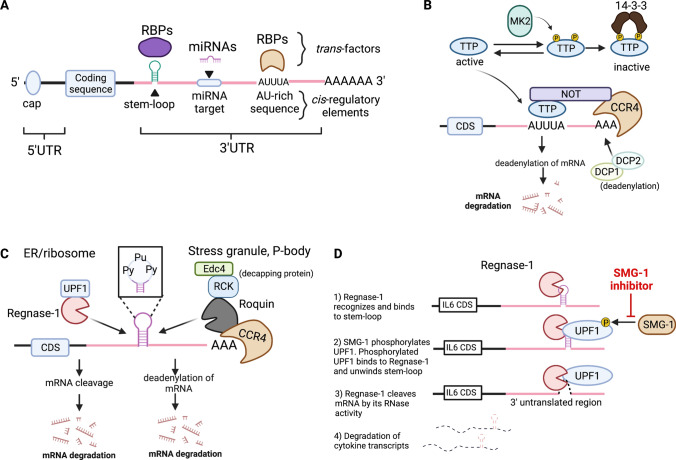
Fig. 2.
Post-transcriptional regulation of inflammatory mRNAs by different RBPs. A Schematic drawing of different cis-regulatory elements in a mRNA. The cis-regulatory elements in the 3’ UTR include stem-loop secondary structure, AU-rich elements and miRNA target sequence. These elements are recognized by RNA-binding proteins or miRNA through cognate RNA-binding domains and distinct sequence, respectively. B TTP recognizes the AU-rich elements in the 3’ UTR of target genes and induces exonucleolytic mRNA decay by recruiting the CCR4-NOT deadenylase complex and DCP1/2 decapping enzymes. Under homeostatic condition, MK2 phosphorylates TTP at two distinct serine residues, and the phosphorylated TTP is sequestered by the 14-3-3 protein, making it inaccessible to the target mRNAs. Upon PRR stimulation, however, its phosphorylation is readily reversible by a protein phosphatase, allowing it to access to the inflammatory cytokine transcripts. C Regnase-1 and Roquin are well-characterized RBPs that recognize stem-loop structures located at the 3’ UTR of common target transcripts. A pyrimidine-purine-pyrimidine (Py-Pu-Py) tri-nucleotide sequence at the stem-loop region preferentially associates with Regnase-1 and Roquin. While Regnase-1 degrades mRNAs with the association of the helicase UPF1 in the ribosome and endoplasmic reticulum (ER), Roquin targets transcripts by recruiting a CCR4-NOT deadenylase complex and Edc4/RCK decapping proteins in the possessing body (PB) and stress granules (SG). CDS, coding sequence. D Sequential dynamics of Regnase-1 degradation of mRNA is mediated by unwinding of stem-loop structures by the helicase UPF1. Small molecule SMG-1 inhibitor blocks SMG1-mediated phosphorylation of UPF1. This figure is made with BioRender: https://biorender.com/