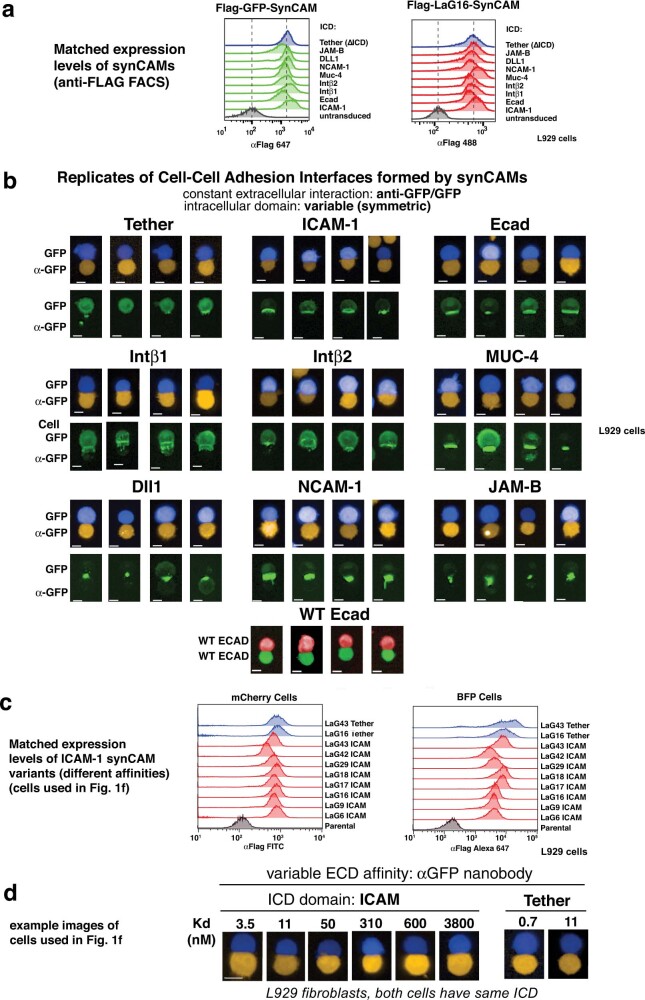
Extended Data Fig. 1. Characterization of synCAM expression and function in L929 fibroblast cells (linked to main Fig. 1c–f).
(a) FACS analysis of GFP (left) and αGFP (right) synCAM expression in L929 fibroblast cells following cell sorting. Surface expression of each synCAM is measured using labelled anti-FLAG tag antibody. The CAM TM and ICD domain for each construct is indicated (tether = control lacking ICD, DLL1 = Delta-like Protein 1, JAM-B = Junction Adhesion Molecule B, NCAM-1 = Neural Cell Adhesion molecule 1, MUC-4 = Mucin 4, ICAM-1 = Intercellular Adhesion Molecule 1, Ecad = E-cadherin, Intβ1 = beta 1 integrin, Intβ2 = beta 2 integrin). Analysis shows that surface expression levels of the tether and alternative synCAM constructs are well matched. (b) Additional replicates of synCAM cell–cell adhesion interface analysis. Maximum projection of 20x confocal microscopy images of pairwise synCAM interfaces (t = 3 h): GFP-expressing cell (blue) is bound to an αGFP expressing cell (orange). The GFP channel of the interfaces is shown, highlighting differences of receptor enrichment. Four out of twenty additional examples are shown here. (c) FACS analysis of αGFP synCAM expression in L929 fibroblast cells expressing cytosolic mCherry (left) or BFP (right) following cell sorting. The CAM TM and ICD domain for each construct is ICAM-1, and the GFP-binding llama nanobody (LaG) ECD for each construct is indicated. This analysis shows that this series of alternative affinity synCAMs are expressed at comparable levels. (d) Maximum projection of 20X confocal microscopy images of pairwise synCAM interfaces (t = 3 h): GFP-expressing cell (blue) is bound to an αGFP expressing cell with the indicated binding Kd (orange).