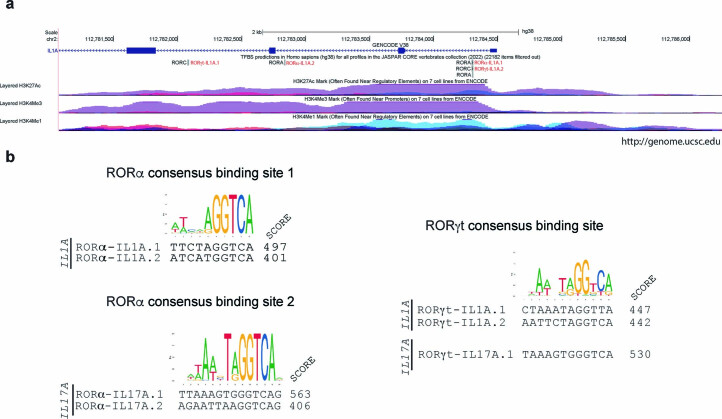
Extended Data Fig. 4. Consensus binding sites for the Th17 master transcription factors ROR-γt and RORα in the IL1A intronic enhancer region.
a, Locations of RORα and ROR-γt consensus binding sites in the regulatory region of IL1A. Binding sites were predicted using the JASPAR2022 database (Castro-Mondragon JA et al., Nucleic Acids Res 2022) and annotated with the UCSC genome browser (assembly hg38) (Kent WJ et al., Genome Res 2002). The quality cutoff was set at 400 (this score indicates a p value of p = 1*10- (score/100)). ENCODE tracks for histone marks were used to indicate the bona fide IL1A regulatory region (chr2:112,781,072-112,786,251) (Rosenbloom KR et al., Nucleic Acids Res 2013). b, Alignment of the RORα and RORγt consensus binding motif with the predicted binding sites in the IL1A enhancer RORα-IL1A.1 (chr2:112784300-112784309), RORα-IL1A.2 (chr2:112782834-112782843); RORγt-IL1A.1 (chr2:112,782,077-112,782,088); RORγt-IL1A.2 (chr2:112,784,298-112,784,309) and established binding sites in the IL17A enhancer RORα-IL17A.1 (chr6:52181117-52181130), RORα-IL17A.2 (chr6:52180979-52180992) and RORγt-IL17A.1 (chr6:52181118-52181129) (Wang X et al., Immunity. 2012). Scores for individual sites are indicated.