Extended Data Fig. 4. Phylogenetic trees of species containing strains found in food.
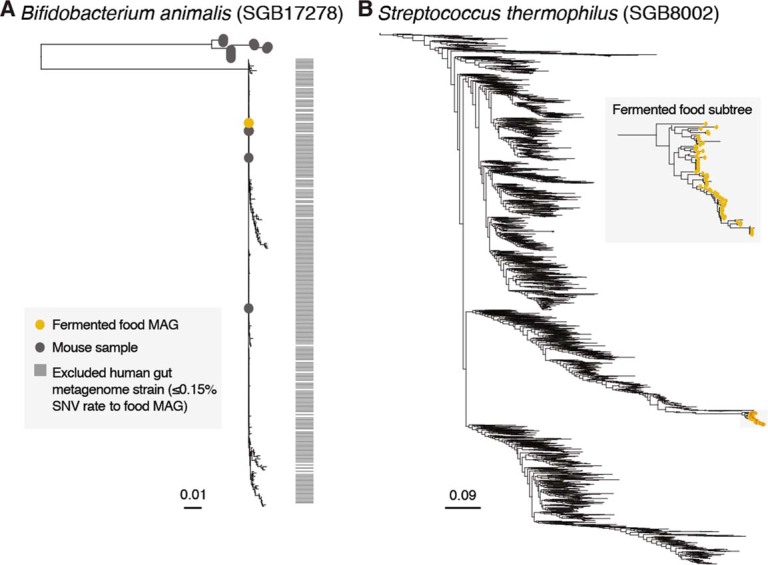
A) Phylogeny of Bifidobacterium animalis (SGB17278) produced with StrainPhlAn (Methods) including strains reconstructed from human gut metagenomes, from mice samples (grey dots) and MAGs reconstructed from fermented food 32 (yellow dots). Differently from strains found in mice, 94% of human-derived strains are at ≤0.0015 single nucleotide variation (SNV) rate to MAGs obtained from fermented food (Methods), suggesting that the presence of this species in humans is associated with consumption of commercial dietary products, and were consequently excluded from further analyses (horizontal grey bars). B) Phylogeny of Streptococcus thermophilus-salivarius-vestibularis (SGB8002) produced with StrainPhlAn (Methods) including strains reconstructed from human gut metagenomes together with MAGs reconstructed from fermented food 32 (yellow dots), suggesting only a subset of strains found in the human gut is associated with fermented food intake. Only the leaves in the enlarged subtree (“Fermented food subtree”) were at ≤0.0015 single nucleotide variation (SNV) rate to MAGs obtained from fermented food (Methods) and were consequently excluded from further analyses.
