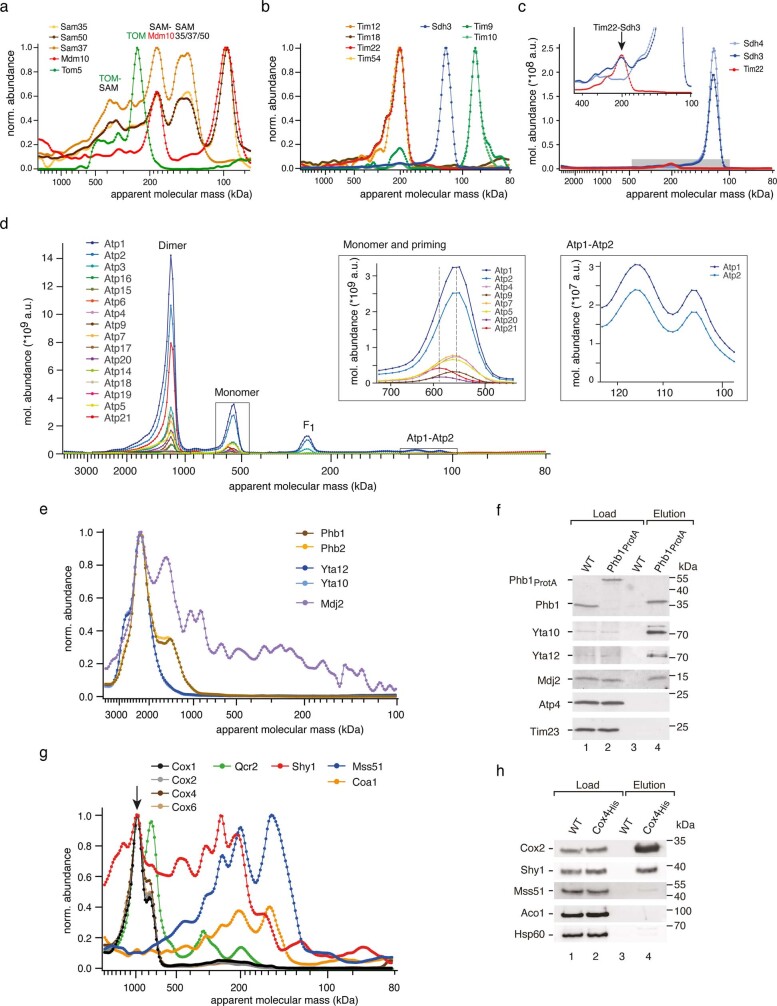
Extended Data Fig. 8. Characterization of dynamic protein machineries by MitCOM.
a, Analysis of the SAM complex illustrates the application of MitCOM in the characterization of dynamic protein machinery. The figure shows normalized abundance–mass profiles of SAM subunits, Mdm10 and Tom5. Distinct forms of SAM and TOM complexes are indicated. The SAM complex, also termed TOB complex for topogenesis of β-barrel proteins, inserts the precursors of β-barrel proteins into the mitochondrial outer membrane. SAM consists of several forms that are in exchange with each other and couple to different partner proteins62,75–79. The SAMcore complex consists of Sam50, Sam37 and Sam3562,65,77. Coupling to TOM (transient TOM-SAM supercomplex) facilitates the transfer of precursor proteins from TOM to SAM, whereas the coupling to Mdm10 (SAM-Mdm10 complex) promotes precursor release62,76,78,80. In addition, Sam37 plays a dynamic role62 and its release from SAMcore leads to a Sam35-Sam50 subcomplex81,82. The apparent overlap of the Sam35-Sam50 subcomplex with Mdm10 in the low molecular mass range does not reflect co-assembly as the peaks can be distinguished by the differences in peak maxima and half-widths. MitCOM efficiently resolves the distinct SAM forms: Sam35-Sam50, SAMcore, SAM-Mdm10, and TOM-SAM supercomplexes. b, c, The interconnected system of the TIM22 complex, the small TIM chaperones Tim9-Tim10, and the SDH complex illustrates a high precision of MitCOM in separation and analysis of complexes of different abundance. The figures show abundance–mass profiles of carrier pathway TIM proteins and SDH (inset in (c): grey box at enlarged scale). The TIM22 complex63,83 migrates in a main peak of ~200 kDa in MitCOM. For Tim9 and Tim10, two populations are resolved, one bound to the TIM22 complex and the other one representing the soluble hetero-hexameric chaperone complex of the intermembrane space. Sdh3 is found in the SDH complex and the ~20-times less abundant TIM22 complex (c-normalized abundance vs. b-absolute abundance)6,63,84. Note that the precursor of Sdh3 is imported and inserted into the inner membrane via the presequence translocase TIM23 and the OXA translocase, not via the TIM22 complex; mature Sdh3 subsequently assembles into the SDH complex and the TIM22 complex85. Sdh4 that is not part of the TIM22 complex does not display a peak at the TIM22 complex (c, insert). MitCOM thus efficiently and quantitatively separates TIM22, Tim9-Tim10 and SDH. d, MitCOM quantitatively separates distinct complexes of the F1FO-ATP synthase. The figure shows full-range abundance profiles of the subunits of F1FO-ATP synthase. The major peak at 1,200 kDa represents the fully assembled dimeric ATP synthase predominating under the mild solubilization conditions used (see Methods), followed by the monomer (at 560 kDa) and the F1-module (at 250 kDa). Left inset, discrimination of two monomeric ATP synthase complex populations. The abundant peak at 560 kDa lacks the dimerizing subunits Atp20 and Atp21. The primed monomeric form at 590 kDa includes Atp20 and Atp2149; note the shoulder-like appearance of the profile peaks of the common subunits caused by the overlap of these two assemblies. Right inset, close co-migration of Atp1 and Atp2 in the low molecular mass range. e, Normalized abundance–mass profiles of prohibitins Phb1 and Phb2, of the m-AAA protease subunits Yta10 and Yta12, and of the inner membrane-bound J-protein Mdj2. The prohibitin 1/2 (Phb1/2) complex forms an oligomeric scaffold and associates with the protease complex m-AAA (Yta10/12)74. In mammalian mitochondria, the J-protein DNAJC19 interacts with the prohibitin complex74, yet it has been unknown if and which yeast mitochondrial J-protein may associate with the prohibitin complex. The inner membrane-bound J-protein Mdj2 migrates in multiple peaks, the largest and most abundant one perfectly co-migrating with the large prohibitin/m-AAA supercomplex of ~2.2 MDa. f, Affinity purification with tagged Phb1 demonstrates that Mdj2 is the yeast J-protein partner of the prohibitin/m-AAA supercomplex. Wild-type (WT) and Phb1ProtA mitochondria were subjected to affinity purification using Protein A sepharose. Bound proteins were eluted with TEV protease and analysed by immunodetection with the indicated antisera. Load 6%, elution 100%. Control proteins: Atp4, subunit of the F1FO-ATP synthase; Tim23, subunit of the presequence translocase of the inner membrane. g, Normalized abundance–mass profiles of structural subunits of respiratory complex IV (Cox1, Cox2, Cox4, Cox6) and complex III (cytochrome bc1 complex; subunit Qcr2), and of the assembly factors Shy1, Coa1 and Mss51. Each of the assembly factors Coa1, Mss51 and Shy1 migrates in multiple peaks in line with their reported presence in various assembly intermediates86–89. A major peak of Shy1 tightly co-migrates with the fully assembled III2IV2 respiratory supercomplex of 1 MDa (arrow), in contrast to Coa1 and Mss51. h, WT and Cox4His mitochondria were subjected to affinity purification via Ni-NTA agarose and analysed by immunodetection with the indicated antisera. Load 2%, elution 100%. Control proteins: Aco1, aconitase; Hsp60, heat shock protein of 60 kDa. Tagged Cox4 that is not part of early Mss51-containing assembly intermediates42 co-purifies Shy1 together with the structural subunit Cox2. Whereas qualitative blue native studies suggested that only a small fraction of Shy1 is associated with the full III2IV2 supercomplex88–90, MitCOM provides a direct quantitative analysis and demonstrates that a main peak of Shy1 is present at the fully assembled supercomplex, different from other assembly factors such as Mss51 and Coa1.