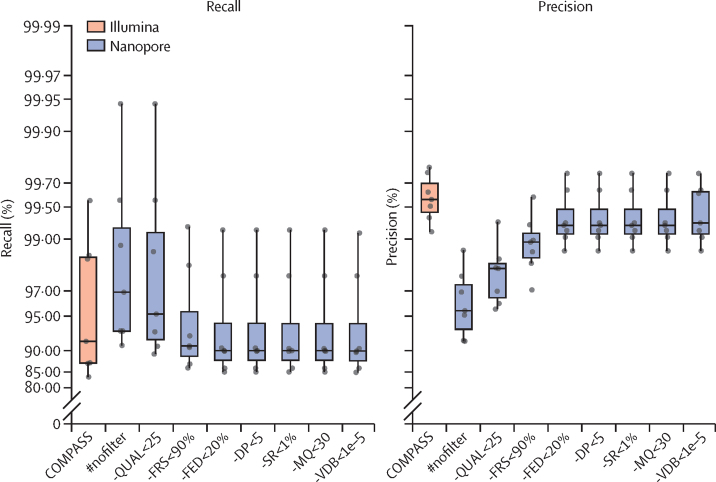
Figure 1.
Recall and precision of SNPs from the Illumina COMPASS pipeline, and the Nanopore BCFtools pipeline with a cumulative selection of filters
Each point represents a single isolate with a PacBio assembly. The midline in each box plot is the median, the upper and lower bounds of each box indicates the span of the quartiles of the data (ie, IQR), and the whiskers extend 1·5 times the IQR. #nofilter is BCFtools with no filtering of variants. Moving right from #nofilter, each box accumulates a new filter plus the previous ones. Each filter describes the criterion for removing an SNP. -QUAL<25 removes SNPs with a quality score less than 25; -FRS<90% removes SNPs whereby less than 90% of reads support the called allele; -FED<20% removes SNPs with read depth below 20% of the isolate's median depth; -DP<5 removes SNPs with less than five reads at the position; -SR<1% removes SNPs with less than 1% of read depth on either strand; -MQ<30 removes SNPs with a mapping quality below 30; -VDB<1e–5 removes SNPs with a variant distance bias less than 0·00001. SNP=single-nucleotide polymorphism.