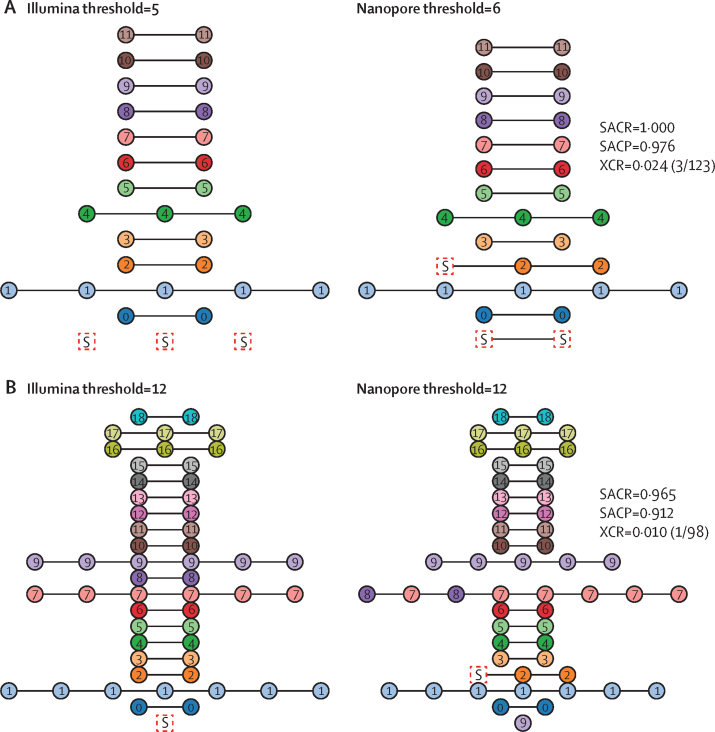
Figure 3.
Agreement of Illumina and Nanopore transmission clustering with thresholds of five for Illumina and six for Nanopore (A) and thresholds of 12 for both technologies (B)
The expected (Illumina COMPASS) clusters are shown on the left and the Nanopore BCFtools clustering is shown on the right. The title of each panel indicates the SNP threshold used for clustering. Nodes are coloured and numbered according to their Illumina cluster membership. Isolates clustered by Nanopore and not clustered (singletons) by Illumina are represented as boxes and are named S. Clusters are horizontally aligned and connected with black lines; however, the order of nodes and the length of edges have no significance. Each Nanopore panel shows the SACR, SACP, and XCR value (with the raw numbers in parentheses) with respect to the Illumina clustering. SACP=sample-averaged cluster precision. SACR=sample-averaged cluster recall. SNP=single-nucleotide polymorphism. XCR=excess clustering rate.