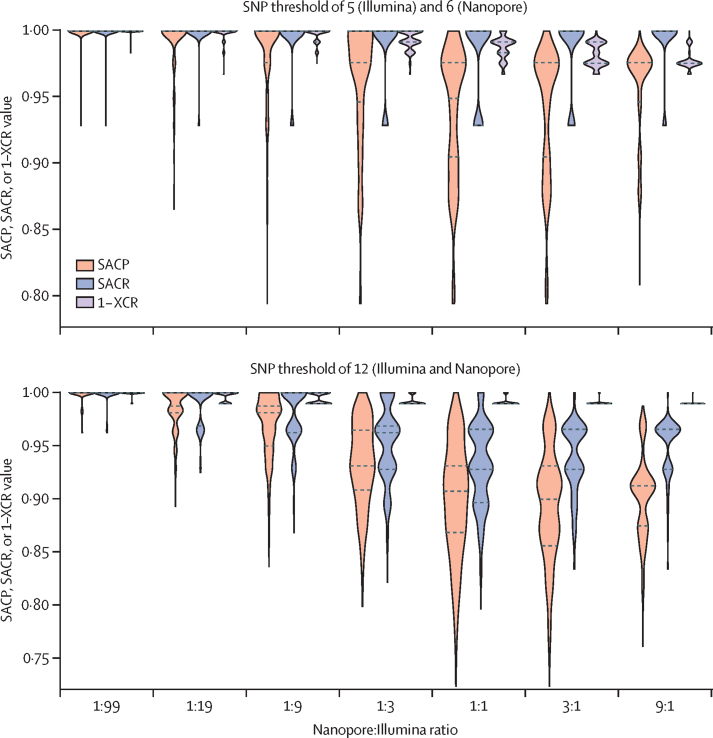
Figure 4.
Simulating heterogeneous datasets with varying proportions of Nanopore and Illumina genomic data
The different thresholds indicate the cutoff for defining isolates as part of a cluster. The y-axis depicts the SACP, SACR, or 1–XCR distributions over all simulation runs. For each ratio and threshold combination we ran 1000 simulations whereby the Nanopore and Illumina data were randomly split into the relevant ratio (eg, 1:9 means one Nanopore isolate for every nine Illumina isolates) and clusters were defined based on the relevant threshold. The titles for each subplot indicate the SNP threshold used when comparing Illumina, Nanopore, or mixed-technology isolate pairs. Dashed horizontal lines show the median and quartiles. SACP=sample-averaged cluster precision. SACR=sample-averaged cluster recall. SNP=single-nucleotide polymorphism. XCR=excess clustering rate.