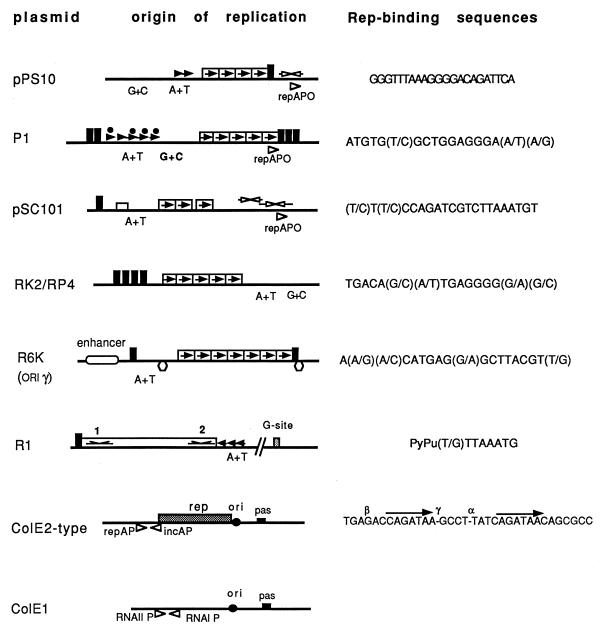
FIG. 1.
Origins of replication and related regions of some representative theta-replicating plasmids from gram-negative bacteria. Plasmid names (left), origins of replication (center), and Rep initiator-binding sites (right) are indicated. The symbols used are as follows: solid boxed arrows correspond to repeated Rep-binding sequences (iterons); open arrows above maps indicate inverted repeats that have partial homology to the iterons; solid arrowheads indicate repeats found in AT-rich regions (A+T); for R1, arrows indicate the imperfect palindromes initially recognized by the RepA initiator protein. Promoters are indicated as open arrowheads below the maps. Other sites of interaction are as follows: IHF-binding sites (open rectangles), dnaA boxes (solid rectangles); FIS-binding sites (hexagons), dam methylation sites (solid circles), and primase assembly sites (pas). Other sites are indicated in the figure. Maps are based on the following references: pPS10 (104, 215); pSC101 (132, 284); P1 (6); RK2 (278); R6K (277); R1 (101); ColE1 (280, 301); ColE2-type plasmids (124).