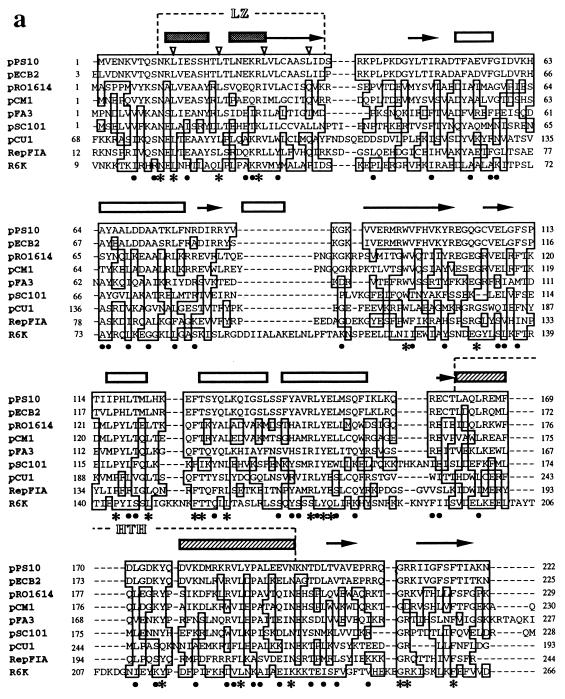
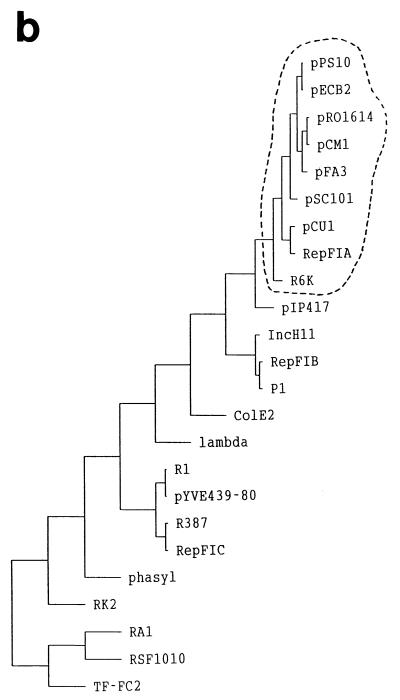
FIG. 2.
Sequence alignment and phylogenetic tree of Rep initiator proteins from theta-replicating plasmids. (a) Sequence alignment of the Rep initiator proteins of nine related plasmids of the iteron-containing class. Sequences were aligned with the program CLUSTAL W (version 1.5) by using, for pairwise alignment, gap opening and extension penalties of 10.0 and 0.1, respectively, and the protein weight matrix BLOSUM30. For multiple alignment, the delay for divergent sequences was set to 40% (294). The degree of sequence identity to the pPS10 initiator sequence for conserved residues is shown: ∗, identical residues in eight or nine of the total sequences; •, identical residues in five to seven sequences. Over the sequences is shown a secondary-structure prediction performed by the neural network algorithm PHD (260) on the output from CLUSTAL W: predicted α-helical regions (boxes) and β-strands (arrows). The two characteristic motifs found in the Rep initiators, LZ (hydrophobic heptad residues pointed to by open arrowheads) and HTH, are indicated. The EMBL database accession numbers are as follows: pPS10, X58896; pECB2, Y10829; pRO1614, L30112; pCM1, X86092; pFA3, M31727; pSC101, K00828; pCU1, Z11775; RepFIA, Y00547; R6K, M65025. (b) Phylogenetic tree for theta-type replicons from gram-negative bacteria, based on sequence alignments of their Rep proteins (such as the one shown in panel a for the pPS10 family, encircled in the tree with a dashed line). The sequences for 35 initiators were retrieved from databases, and a preliminary alignment was performed with CLUSTAL W (data not shown). Sequences that were virtually identical (pairwise scores, ≥90) were discarded, and a refined alignment was used as input data for the DISTANCES program of the Genetics Computer Group software package (95). Distance matrixes were corrected for multiple substitutions by the method of Kimura. The phylogenetic tree was built up according to the UPGMA method with the program GROWTREE (95). For further discussion, see the text.