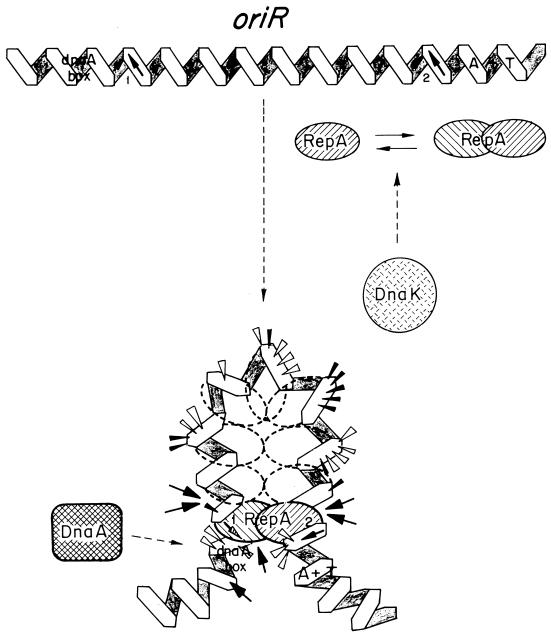
FIG. 4.
RepA-oriR complexes in initiation of R1 plasmid replication. A 100-bp region in the oriR replication origin is continuously bound by the plasmid RepA protein to form the initiation complex. There are no iterons in oriR, but two partially palindromic 10-bp sequences (sites 1 and 2) are found at the ends of that 100-bp region. They are flanked, respectively, by a consensus dnaA box and by three AT-rich sequences. These three sequences are believed to be melted to allow the DnaBC complex access to the open origin. A RepA initiator (hatched oval) dimer binds with high affinity to site 1, and then, in a second binding event, a different RepA dimer would bind with lower affinity to the distal site 2 sequence. The DNA of the oriR region could be bent to facilitate the topological proximity of sites 1 and 2, which are disposed on the same face of DNA double helix. Binding of RepA dimers to sites 1 and 2 would generate a small DNA loop, held together by protein-protein interactions. The DNA loop would be filled afterwards with more RepA molecules that are brought to the complex mainly by protein-protein interactions. This model is based on experimental gel mobility shift assays and footprinting data with both wild-type and mutant RepA proteins and oriR sequences (101). Arrowheads indicate DNase I-hypersensitive sites (the size is proportional to the intensity of cleavage), whereas arrows point to strong cleavage sites for hydroxyl radicals. The interaction of DnaA host initiator with its DNA-binding site is dependent on the previous formation of the RepA-oriR nucleoprotein complex (233). A requirement for a DnaK chaperone has been described for R1 DNA replication (105). A hypothetical role for DnaK in modulating the aggregation and activation state of RepA dimers, inspired by the findings for P1 plasmid RepA initiator (316), is also shown.