Abstract
Background
In today’s society, cancer has become a big concern. The most common cancers in women are breast cancer (BC), endometrial cancer (EC), ovarian cancer (OC), and cervical cancer (CC). CC is a type of cervix cancer that is the fourth most common cancer in women and the fourth major cause of death.
Results
This research uses a network approach to discover genetic connections, functional enrichment, pathways analysis, microRNAs transcription factors (miRNA-TF) co-regulatory network, gene-disease associations, and therapeutic targets for CC. Three datasets from the NCBI’s GEO collection were considered for this investigation. Then, using a comparison approach between the datasets, 315 common DEGs were discovered. The PPI network was built using a variety of combinatorial statistical approaches and bioinformatics tools, and the PPI network was then utilized to identify hub genes and critical modules.
Conclusion
Furthermore, we discovered that CC has specific similar links with the progression of different tumors using Gene Ontology terminology and pathway analysis. Transcription factors-gene linkages, gene-disease correlations, and the miRNA-TF co-regulatory network were revealed to have functional enrichments. We believe the candidate drugs identified in this study could be effective for advanced CC treatment.
Background
In 2020, an estimated 19.3 million new cancer cases (18.1 million excluding nonmelanoma skin cancer) were diagnosed worldwide, with around 10.0 million cancer deaths (9.9 million excluding nonmelanoma skin cancer) [1]. The global cancer burden is expected to be 28.4 million cases in 2040, a 47% increase from 2020, with a greater increase in transitioning (64 to 95%) countries versus transitioned (32 to 56%) countries due to demographic changes, though this may be exacerbated further by increasing risk factors associated with globalization and a growing economy [2, 3]. Lung cancer and colorectal cancer are the most prevalent cancers in men, whereas breast cancer, colorectal cancer, cervical cancer, and lung cancer are the most common in females [4–6]. In 2020, men had a 19% higher overall cancer incidence rate (222.0 per 100,000) than women (186 per 100,000), while rates varied greatly across areas. Cervical cancer is the fourth most common malignancy in women and the fourth greatest cause of cancer mortality, with an estimated 604,000 new cases and 342,000 deaths in 2020 [7]. The major risk factor for CC is infection with particular forms of HPV, followed by smoking [1, 8]. Types of HPV are 16 and 18 accounting for 75% of all CC instances globally, with HPV types 31 and 45 accounting for the remaining 10% [9]. There are approximately 150–200 different types of HPV that have been identified, with 15 being considered high risk (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82), three being considered probable high risk (26, 53, and 66), and 12 being considered low risk (6, 11, 40, 42, 43, 44, 54, 61, 70, 72, 81, and CP6108) [10].
An investigation of differentially expressed gene (DEG) promoter sequences and transcription factor (TF) binding sites previously identified TF E2F as a crucial transcriptional regulator and a possible molecular target for cervical cancer treatment [11]. Another research found that the target genes CDC45, GINS2, MCM2, and PCNA are important participants of cervical cancer [12]. Several novel candidate genes implicated in cervical carcinogenesis (e.g., VEGFA and IL-6) were also predicted by combining human protein interaction data with cervical cancer gene sets [13, 14]. These researches have provided important information concerning cervical cancer, but no conclusions about the disease’s underlying molecular pathways were made. Another wisely analyzed study has demonstrated that the predictive potential reporter biomolecules including KAT2B, PCNA, CD86 [15, 16], miR-192-5p, and miR-215-5p have a significant role in cervical cancer [17].
Three datasets have been taken to complete this network-based study. The common DEGs were then determined by comparing the datasets. Using common DEGs have applied for GO annotation, pathway analysis, construct PPI network, module analysis, hub DEGs identification, gene-disease association prediction, TF-miRNA co-regulatory network identification, and drug target prediction.
Methods
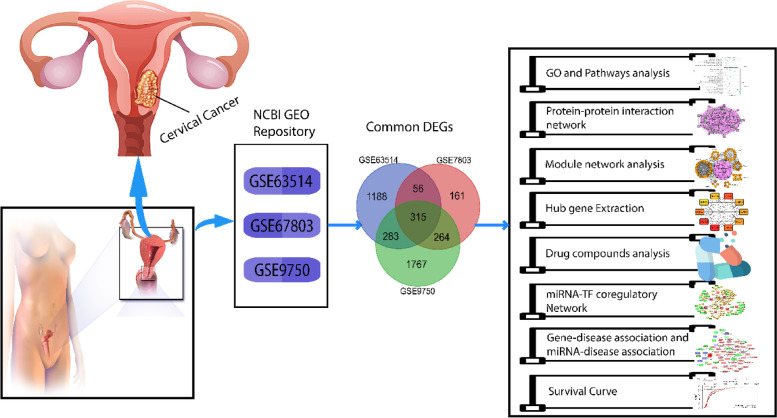
The study effort has included differential expressed genes (DEGs) discovery, common DEGs finding, gene ontology analysis, pathways enrichment analysis, protein interaction network building, module analysis, TF-miRNA co-regulatory network, drug compounds analysis, and gene-disease correlations network creation, which is a network-based approach. All methodology phases are detailed here, and a quick representation of this work is demonstrated in Fig. 1.
Fig. 1.
Image workflow of this study methods. A transcriptomic comparative analysis was performed for CC. Three datasets were gathered for CC, and GEO2R was used to detect differentially expressed genes (DEGs), and the datasets were filtered to normalize and find common DEGs. Using shared datasets has performed various transcriptomic analyses
Data collection
The GEO database is a free public resource that stores and distributes high-throughput gene expression data from all over the world (https://www.ncbi.nlm.nih.gov/geo/) [18–20]. Three datasets have been considered for CC for this study including GSE9750, GSE63514, and GSE7803 from the GEO repository. GSE9750 stands on a single platform GPL96 [HG-U133A] Affymetrix Human Genome U133A Array for Homo sapiens organisms named “Identification of gene expression profiles in CC.” This dataset conducts total of 66 samples where 42 samples are affected and 24 samples are normal cervical cell [21]. GSE63514 dataset depends on a single platform GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array. This dataset conducts a total of 128 samples where 104 are affected cells and 24 normal cells [22]. And the last dataset GSE7803 conducts 10 normal cells and 31 affected cells [23].
DEGs identification
GEO2R was utilized to find the DEGs. The raw file was transformed to expression using a statistical relevance p-value of 0.05 and logFc > 1 (upregulated) and logFc-1 (downregulated), and the common DEGs were identified using a Venn diagram web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/) [24].
Ontological terms and pathways analysis
The biological activities of the common DEGs were identified via gene ontology analysis using the Gene Ontology database [25], while the corresponding pathways were discovered in the KEGG database [26]. The Enrichr online tool [27] was used for all of the analyses. Enrichr is a web-based intuitive enrichment analysis application that provides many sorts of graphical summaries of gene list collective functions. Enrichr is an open-source project that can be found at http://amp.pharm.mssm.edu/Enrichr [28].
Construction of PPI network and clustering analysis
The PPI network [29] is a graphical representation of gene interaction. The PPI network was built using the physical interactions of proteins from frequent DEGs in CC datasets from the STRING database [30]. To predict the interconnection between the proteins was set the minimum confidence score of 0.40. Afterward, the open-source Cytoscape tool was used to analyze the PPI network [31]. To identify the complex network part (cluster) of the PPI network, the MCODE method was implemented [32]. The MCODE basic parameters degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and maximum depth = 100 were chosen as a minimum criterion. Using the connection (degree) technique, the cytoHubba plugin program of Cytoscape software was used to discover hub DEGs from the PPI network [33].
Drug target analysis
The drug signatures database (DSigDB) drug signature database [34] was used to identify therapeutic targets for selected hub DEGs. The DSigDB is a new gene set resource for gene set enrichment analysis that links drugs/compounds to their target genes (GSEA). DSigDB now has 22,527 gene sets, each of which contains 17,389 unique compounds spanning 19,531 genes. Users may search, browse, and download drugs, chemicals, and gene sets from the DSigDB database. In GSEA software, DSigDB gene sets may be utilized to connect gene expression to drugs/compounds for drug repurposing and translational research [34]. The cutoff criterion for identifying pharmacological targets was p-value 0.01 and overlapping genes count > = 9.
Association network of gene disease
Linkage studies [35], genome-wide association studies (GWAS) [36], and RNA interference screens [37] are all expensive and time-consuming approaches for determining gene-disease associations. As a result, a variety of computational methods [38–40] for identifying or predicting gene-disease correlations have been developed. These approaches have distinct strengths and limitations, and they are best suited to different types of diseases [41]. Gene-disease association network has been exported through DisGeNET database (https://www.disgenet.org/).
TF-miRNA co-regulatory network development and analysis
The RegNetwork repository (http://www.regnetworkweb.org/) was used to build the TF-miRNA co-regulatory networks that revealed new information about the key role of proposed TF-miRNA co-regulation in CC and its crosstalk with the surrounding microenvironment [42]. RegNetwork is a human and mouse gene regulatory network repository that collects and combines known regulatory connections between TFs, miRNAs, and target genes from 25 different datasets. RegNetwork is a database that houses a comprehensive collection of empirically known or predicted transcriptional and posttranscriptional regulatory interactions, and the database structure is flexible enough to accommodate future additions into gene regulatory networks for more species [43].
Analysis of survival for hub genes
A time-to-event study, also known as a survival analysis, refers to a set of approaches for long-term research prior to the identification of a specific endpoint of interest. The occurrence (e.g., death) does not frequently occur in all patients at the end of the observation period [44–46], which is a distinctive feature of the survival data. In this analysis, the survival role was calculated for altered and regular classes of significant genes common to datasets of CC by using Cox PH and PL estimators. The Cox PH regression model was evaluated univariate as well as multivariate [47].
Results
Three-hundred fifteen common DEGs identified
Using GEO2R for CC, the dataset was downloaded from the GEO repository of NCBI. Initially, the GSE9750, GSE63514, and GSE7803 datasets revealed 21,156, 45,118, and 20,056 DEGs, respectively, after filtration with the criteria 2629, 1842, and 796 DEGs were taken. Following that, a comparison of the DEGs revealed 315 shared DEGs (Fig. 2).
Fig. 2.
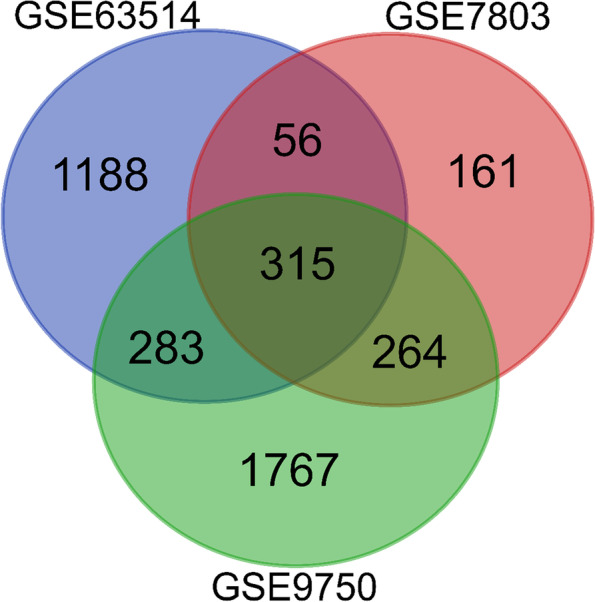
A comparison method was applied between the datasets including GSE63514, GSE7803, and GSE9750 to identify the common DEGs. A total of 315 common DEGs were identified for further analysis
Ontological terms and pathways analysis
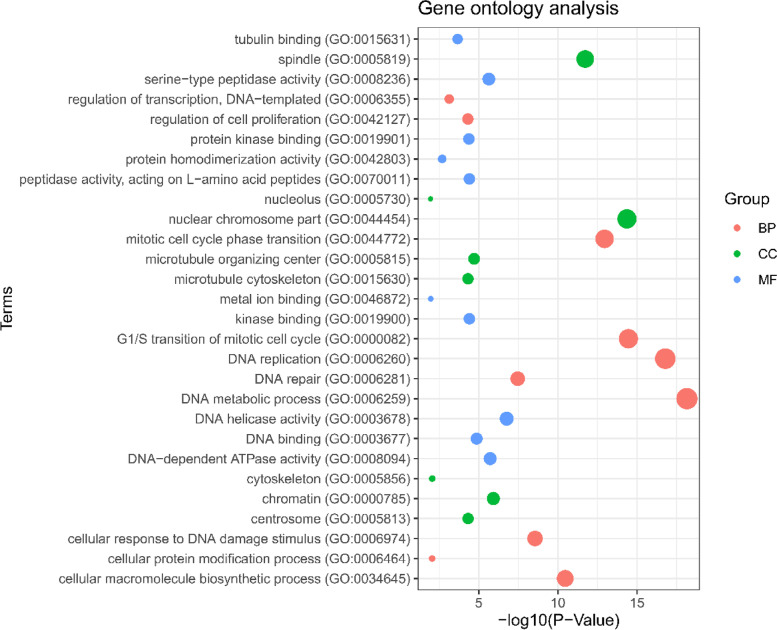
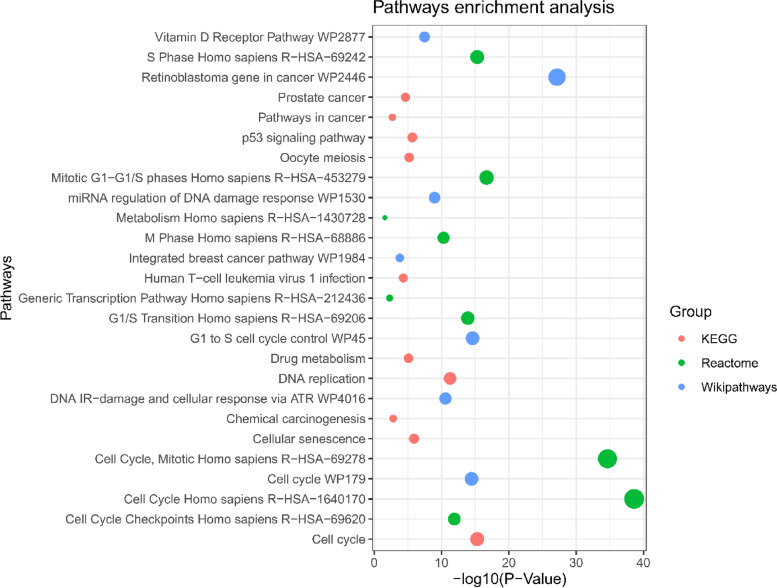
The GO analysis reveals most of the DEGs associated with the terms including the following: DNA metabolic process, DNA replication, G1/S transition of mitotic cell cycle, mitotic cell cycle phase transition, cellular macromolecule biosynthetic process, cellular response to DNA damage stimulus, DNA repair, regulation of cell proliferation, regulation of transcription, DNA templated, cellular protein modification process for biological process, nuclear chromosome part, spindle, chromatin, microtubule-organizing center, centrosome, microtubule cytoskeleton, cytoskeleton, nucleolus for cellular component, DNA helicase activity, DNA-dependent ATPase activity, serine-type peptidase activity, DNA binding, peptidase activity, acting on L-amino acid peptides, kinase binding, protein kinase binding, tubulin binding, protein homodimerization activity, metal ion binding for molecular function, etc. (Table 1) (Fig. 3). The pathway of KEGG, Reactome, and WikiPathways analysis reveals that most of the common DEGs were associated with the cell cycle, DNA replication, cellular senescence, cell cycle, p53 signaling pathway, pathways in cancer, mitotic, mitotic G1-G1/S phases, S phase, M phase, generic transcription pathway, prostate cancer, metabolism, retinoblastoma gene in cancer, G1 to S cell cycle control, integrated breast cancer pathway, etc. (Table 2) (Fig. 4).
Table 1.
Significant GO terms with their GO ID, overlapped genes, and p-value of the common DEGs associated with CC. In this table, BP means biological process, MF means molecular function, and CC means cellular component. All the GO terms which have been taken were filtered with a p-value less than 0.05. Basically, the p-value helps to determine the significance of the results in relation to the null hypothesis
| Terms | GO ID | p-value | Overlapped genes | Group |
|---|---|---|---|---|
| DNA metabolic process | GO:0006259 | 7.16E-19 | 34 | BP |
| DNA replication | GO:0006260 | 1.65E-17 | 22 | BP |
| G1/S transition of mitotic cell cycle | GO:0000082 | 3.52E-15 | 19 | BP |
| Mitotic cell cycle phase transition | GO:0044772 | 1.13E-13 | 24 | BP |
| Cellular macromolecule biosynthetic process | GO:0034645 | 3.48E-11 | 27 | BP |
| Cellular response to DNA damage stimulus | GO:0006974 | 2.79E-09 | 23 | BP |
| DNA repair | GO:0006281 | 3.47E-08 | 20 | BP |
| Regulation of cell proliferation | GO:0042127 | 4.85E-05 | 27 | BP |
| Regulation of transcription, DNA templated | GO:0006355 | 7.29E-04 | 42 | BP |
| Cellular protein modification process | GO:0006464 | 0.008786 | 26 | BP |
| Nuclear chromosome part | GO:0044454 | 4.36E-15 | 33 | CC |
| Spindle | GO:0005819 | 1.90E-12 | 21 | CC |
| Chromatin | GO:0000785 | 1.19E-06 | 18 | CC |
| Microtubule-organizing center | GO:0005815 | 1.96E-05 | 22 | CC |
| Centrosome | GO:0005813 | 4.70E-05 | 20 | CC |
| Microtubule cytoskeleton | GO:0015630 | 4.78E-05 | 18 | CC |
| Cytoskeleton | GO:0005856 | 0.008643 | 16 | CC |
| Nucleolus | GO:0005730 | 0.011101 | 19 | CC |
| Serine-type peptidase activity | GO:0008236 | 2.30E-06 | 15 | MF |
| DNA binding | GO:0003677 | 1.36E-05 | 32 | MF |
| Peptidase activity, acting on L-amino acid peptides | GO:0070011 | 3.89E-05 | 12 | MF |
| Kinase binding | GO:0019900 | 3.90E-05 | 19 | MF |
| Protein kinase binding | GO:0019901 | 4.20E-05 | 21 | MF |
| Tubulin binding | GO:0015631 | 2.16E-04 | 13 | MF |
Fig. 3.
Gene ontological analysis for BP, CC, and MF. Red-colored terms indicate BP, green-colored terms indicate CC, and blue-colored terms indicate MF. According to figure, DNA metabolic process (GO: 0006259) is the most significant term from the BP ontology
Table 2.
Pathways enrichment analysis with their p-value and overlapped genes of common DEGs for CC through three databases including KEGG, Reactome, and WikiPathways. The pathways of the cell cycle are the most significant which have been reported by the three databases
| Pathways | p-value | Overlapped genes | Database |
|---|---|---|---|
| Cell cycle | 4.92E-16 | 21 | KEGG |
| DNA replication | 5.26E-12 | 11 | KEGG |
| Cellular senescence | 1.19E-06 | 13 | KEGG |
| p53 signaling pathway | 2.13E-06 | 9 | KEGG |
| Oocyte meiosis | 6.35E-06 | 11 | KEGG |
| Drug metabolism | 8.07E-06 | 10 | KEGG |
| Prostate cancer | 2.26E-05 | 9 | KEGG |
| Human T-cell leukemia virus 1 infection | 4.70E-05 | 13 | KEGG |
| Chemical carcinogenesis | 0.001498 | 11 | KEGG |
| Pathways in cancer | 0.002009 | 18 | KEGG |
| Cell cycle | 2.40E-39 | 67 | Reactome |
| Cell cycle, mitotic | 2.20E-35 | 58 | Reactome |
| Mitotic G1-G1/S phases | 1.97E-17 | 23 | Reactome |
| S phase | 4.92E-16 | 21 | Reactome |
| G1/S transition | 1.22E-14 | 19 | Reactome |
| Cell cycle checkpoints | 1.24E-12 | 21 | Reactome |
| M phase | 4.93E-11 | 23 | Reactome |
| Generic transcription pathway | 0.005002 | 23 | Reactome |
| Metabolism | 0.025365 | 41 | Reactome |
| Retinoblastoma gene in cancer | 6.87E-28 | 27 | WikiPathways |
| G1 to S cell cycle control | 2.42E-15 | 16 | WikiPathways |
| Cell cycle | 3.43E-15 | 20 | WikiPathways |
| DNA IR-damage and cellular response via ATR | 2.57E-11 | 14 | WikiPathways |
| miRNA regulation of DNA damage response | 1.05E-09 | 12 | WikiPathways |
| Vitamin D receptor pathway | 3.27E-08 | 16 | WikiPathways |
| Integrated breast cancer pathway | 1.52E-04 | 10 | WikiPathways |
Fig. 4.
Pathways enrichment analysis through three databases including KEGG, Reactome, and WikiPathways. All the revealed pathways had been performed p-value less than 0.05
Hub genes identification and clustering analysis
The PPI network is a significant product of this sort of research. The STRING database was used to create the PPI network, which was then shown using the Cytoscape application. The PPI network (Fig. 5A) has 183 nodes and 1082 edges, with nodes representing DEGs and edges representing connections between nodes. Afterward, we have gotten 10 hub DEGs (CDK1, CCNB1, CDC20, TOP2A, MAD2L1, NDC80, AURKA, ASPM, NCAPG, and BIRC5) (Fig. 5B) using cytoHubba plugin tool of Cytoscape. By contrast, the MCODE plugin algorithm showed 10 modules (cluster sub-networks); from them, significant 7 modules have taken (Fig. 6). The first module carried 27 nodes and 316 edges, the second module contains 9 nodes and 36 edges, the third module conducts 15 nodes and 51 edges, and fourth modules carried 10 nodes and 26 edges.
Fig. 5.
A Protein interaction for the common differentially expressed genes for cervical cancer. The network conducts 183 nodes (indicating genes) and 1082 edges (indicating connections between nodes). B The hub genes were identified from the protein interaction network. Top 10 significant genes were CDK1, CCNB1, CDC20, TOP2A, MAD2L1, NDC80, AURKA, ASPM, NCAPG, and BIRC5
Fig. 6.
Seven significant modules could be extracted through the MCODE plugin tools of Cytoscape. The module is the very complex area of PPI network. In the figure, yellow color circular nodes represent the module network. There were seven module networks which have been obtained
Lucanthone and paclitaxel target significantly interacted with the hub genes
The therapeutic target for the hub DEGs was discovered using the DSigDB drug target database. The result showed that the “LUCANTHONE CTD 00006227 and paclitaxel CTD 00007144” both of drug targets mostly associated with the hub DEGs (Table 3). Lucanthone is being studied as a chemotherapy and radiation sensitizer due to its potential to interact with DNA repair [48]. However, because lucanthone reduces cancer cell survival regardless of p53 status, autophagy suppression may be a more important contribution to the lucanthone mode of action that impacts DNA repair. Because lucanthone suppresses autophagy, it may be able to boost the effectiveness of chemotherapeutics that activate this process. Both apoptosis and autophagy have been observed to be induced by vorinostat and bortezomib, and inhibiting autophagy increases its activity [49–51]. Tumor immunotherapy improves the body’s immunity, resulting in the immunological response to malignancies. Our understanding of the potential utility of conventional medicines in tumor immunotherapy has advanced recently [52]. According to multiple studies, paclitaxel directly eliminates tumor cells while also regulating immune cells such as effector T cells, dendritic cells (DCS), natural killer (NK) cells, regulatory T cells (Tregs), and macrophages [53]. Belinostat [54], doxorubicin [55], bleomycin [56], and bortezomib [57] are examples of chemotherapeutics with comparable immunomodulatory characteristics. Paclitaxel is a novel anticancer medication having broad-spectrum action in epithelial ovarian cancer, head and neck cancer, esophageal cancer, breast cancer, CC, and lung cancer [58–60]. The Food and Drug Administration (FDA) has approved cisplatin and topotecan for the treatment of advanced cervical cancer. However, the cisplatin/paclitaxel or carboplatin/paclitaxel regimens are less toxic and easier to administer than cisplatin/topotecan according to National Comprehensive Cancer Network (NCCN) Guidelines Insights: Cervical Cancer, Version 1.2020 [61, 62]. The panel decided to add carboplatin/paclitaxel/bevacizumab to the list of recommended regimens for recurrent or metastatic cervical cancer based on the findings of GOG 240 and JGOG0505. According to prior research, cisplatin/paclitaxel and carboplatin/paclitaxel have become the most frequently utilized systemic regimens for metastatic or recurrent cervical cancer [61].
Table 3.
Significant drug compounds with overlapped DEGs and p-value for hub DEGs. From the analysis of drug compounds, lucanthone and paclitaxel showed the high significance for the CC. Lucanthone and paclitaxel overlapped 10 and 9 hub DEGs respectively
| Drug compounds | Overlapped genes | p-value |
|---|---|---|
| LUCANTHONE CTD 00006227 | 10 | 1.52E-20 |
| Paclitaxel CTD 00007144 | 9 | 3.06E-14 |
| Dasatinib CTD 00004330 | 9 | 3.60E-14 |
| Troglitazone CTD 00002415 | 9 | 3.77E-13 |
| Resveratrol MCF7 DOWN | 6 | 3.53E-12 |
| Dasatinib CTD 00004330 | 9 | 3.60E-14 |
Gene-disease association
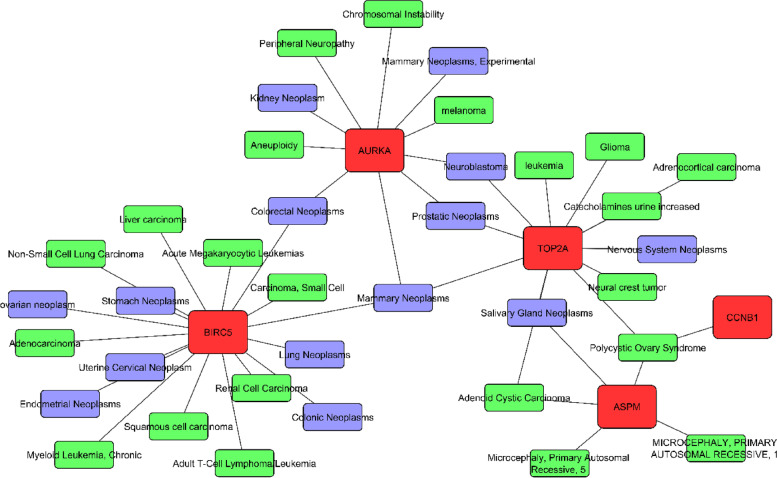
Hub genes cooperated to explore the gene-disease interaction network through the DisGeNET database. In the gene-disease interaction, network genes and diseases are interconnected. A total of 10 hub genes were used to apply for the gene-disease association network where just 5 hub genes show the connectivity between the genes and diseases (Fig. 7). The network showed the BIRC5 gene is the most connected node that is associated with 17 cancers and tumor-related diseases.
Fig. 7.
The hub DEGs linked diseases as illustrated by the gene-disease association network. Red nodes, hub genes; green, cancer-related genes; blue, neoplasms
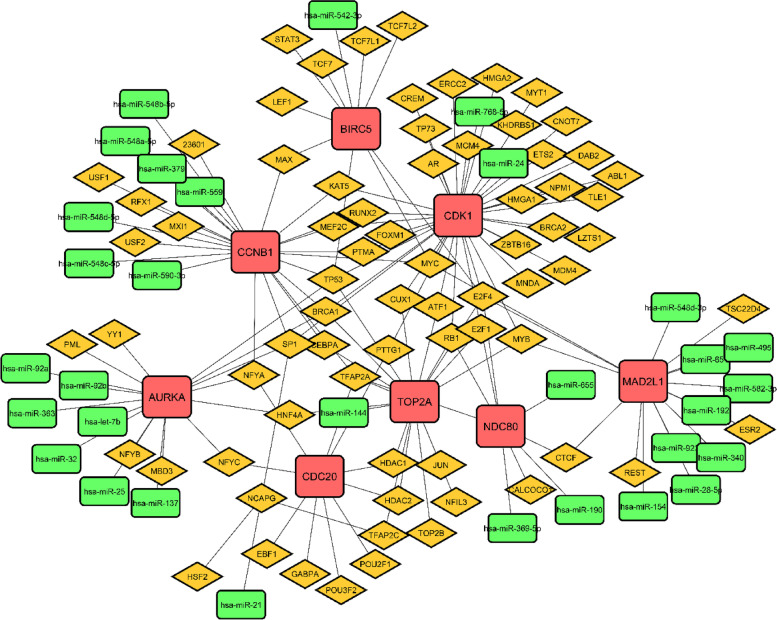
TF-miRNA co-regulatory network development and analysis
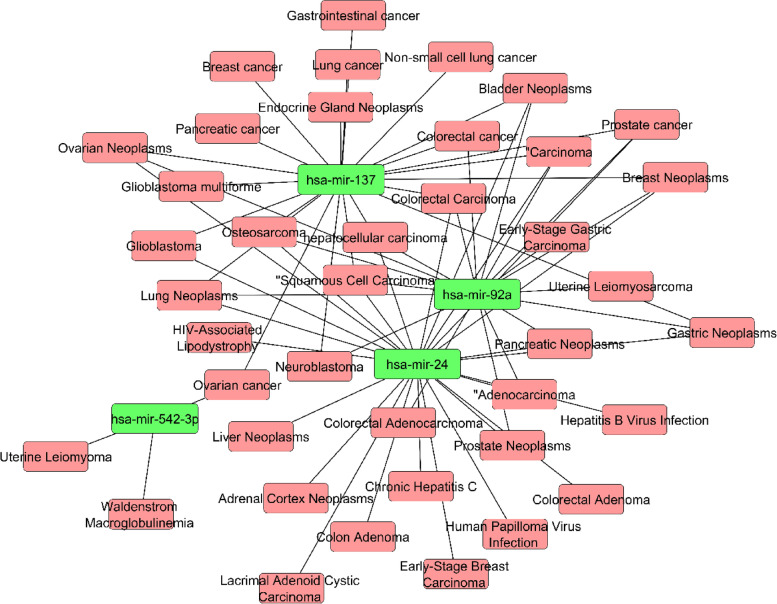
Transcription factors (TFs) and microRNAs (miRNAs) are important regulators of gene expression [63]. In CC, miRNAs and TFs may perform a dual regulatory role. We built a complete particular TF-miRNA co-regulatory network by merging predicted and empirically confirmed TF and miRNA targets after gathering hub genes from the PPI network. Using hub genes, the RegNetwork repository was utilized to build a TF-miRNA co-regulatory network. There are 112 nodes and 136 edges in the TF-miRNA co-regulatory network, including 73 TF candidates, 8 hub nodes, and 31 miRNA candidate nodes (Fig. 8). In addition, 31 miRNA were analyzed to detect the cancer disease connectivity (Fig. 9). In the figure, hsa-mir-137, hsa-mir-92a, and hsa-mir-542-3p show the high connectivity with the disease including breast cancer, ovarian cancer, pancreatic cancer, glioblastoma, non-small cell lung cancer, and bladder neoplasms.
Fig. 8.
In this network, the TF genes and miRNA are interconnected named TF-miRNA co-regulatory network. Herein, the square shape red color nodes indicate hub DEGs, yellow color TF candidates shown as a diamond shape nodes, and rectangular green color nodes indicate miRNA candidates
Fig. 9.
miRNA disease association network showing the interconnection between the miRNA (green color) and disease (red color)
Survival analysis
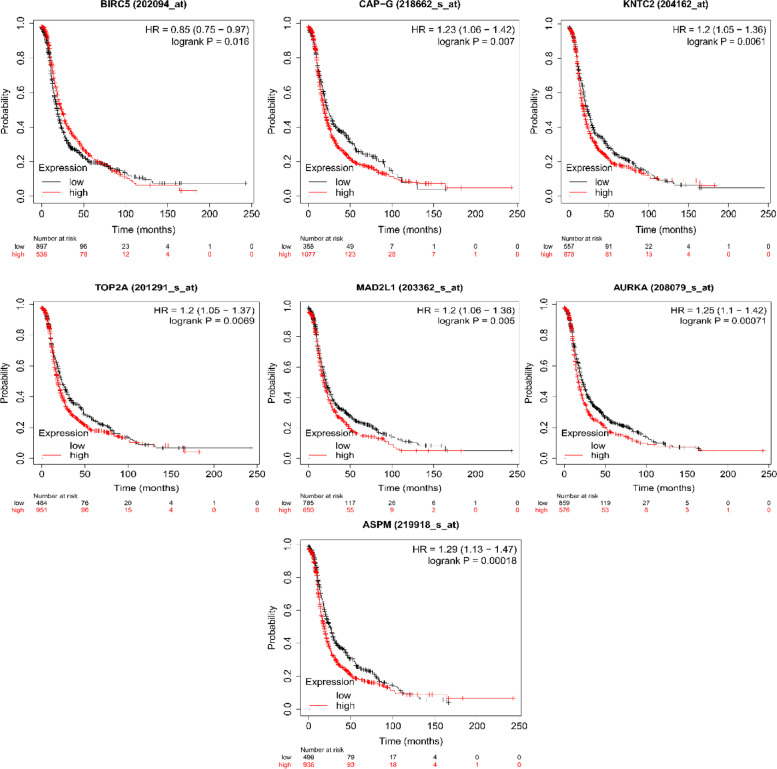
Following the analysis, were picked are the most critical genes of CC with the cutoff p-value <= 0.05. The PL estimator had been used to achieve the survival curves of the most relevant genes in contrast with altered and regular populations. All the hub genes were analyzed for survival rating, and seven proteins including BIRC5, NCAPG/CAPG, TOP2A, MAD2L1, AURKA, ASPM, and NDC80/KNTC2 perform logrank P less than 0.05 (Fig. 10). CAPG could play a significant role in the survival of breast cancer [64], bladder cancer [65], as well as ovarian cancer [66], and many more. This finding reports for the first time that CAPG may play a significant role in the survival of cervical cancer. Although MAD2L1 [67], TOP2A [68], AURKA [69], and ASPM [68] were reported in various research findings, they may play a remarkable role in the survival of cervical cancer. By contrast, NDC80 and CAPG are a novel targets in the survival of cervical cancer.
Fig. 10.
This figure showed the survival rating of patient-associated CC with the common proteins. The survival plot has taken with the logrank p < 0.05. The survival plot is made with two axis named probability and time (months). All the hub proteins were analyzed for survival rating, and seven proteins including BIRC5, NCAPG/CAPG, TOP2A, MAD2L1, AURKA, ASPM, and NDC80/KNTC2 perform less than 0.05
Discussion
CC is the fourth most frequent cancer in women worldwide and the fourth leading cause of cancer death. According to estimations, CC claimed the lives of 342,000 individuals in 2020. Around 8% of all cancer diagnoses and deaths are caused by this condition. We used a network-based technique to look at the patterns of gene expression in three microarray datasets of CC patients and found molecular targets that could be employed as cancer biomarkers. It could also disclose crucial information about their impact on the progression of illnesses or disorders. In the fields of biomedical and computational biology, expression profiling utilizing high-throughput microarray datasets has proven to be a helpful resource for identifying biomarker candidates for a variety of diseases [70]. According to the CC transcriptomics analysis, the common 315 DEGs have comparable expression in three datasets. To obtain insight into the etiology of CC, the biological relevance of 315 frequently DEGs was examined utilizing gene ontology and pathway analysis techniques based on p-values.
The Gene Ontology (GO) is a gene regulation framework based on a generic conceptual perspective that makes it easier to comprehend genes and their interactions. Evolution achieved this over time by accumulating biological knowledge about gene functions and regulation in a variety of ontological domains [71]. For three types of GO analysis, the GO database was employed as an annotation source: BP (molecular activities), CC (gene controls function), and MF (activities at the molecular level) [72]. The biological process (34 genes) and DNA metabolic process (34 genes) are the most important, followed by transcription control (43 genes). Many cellular metabolic activities involve deoxyribonucleic acid. This is one of the two primary types of nucleic acid, and it is made up of one or two strands of connected deoxyribonucleotides [73]. The method by which a cell controls the translation of DNA to RNA (transcription) to regulate gene activity is called transcriptional regulation. Changing the quantity of copies of RNA produced and manipulating when the gene is transcribed are two examples of how a single gene can be managed [74].
Most of the gene promoters in invertebrates have a CpG island with several CpG sites [75]. A gene is silenced when several of its promoter CpG sites are methylation [76]. However, transcriptional silencing may have a bigger role in cancer development than mutation. For the cellular component function, the nuclear chromosome part (33 genes), microtubule-organizing center (22 genes), and chromatin (18 genes) are significant. According to the molecular function, top GO terms peptidase activity, acting on L-amino acid peptides (12 genes), kinase binding (19 genes), and DNA binding (32 genes), are significantly associated.
Pathway analysis [77] is the most effective method for reflecting an organism’s behavior via internal changes. The pathways of the most prevalent DEGs were culled from three separate databases: KEGG, Reactome, and WikiPathways. Cell cycle, DNA replication, cellular senescence, the p53 signaling route, drug metabolism, prostate cancer, human T-cell leukemia virus 1 infection, chemical carcinogenesis, and cancer pathways are the top ten KEGG pathways. Cell cycle, mitotic, mitotic G1-G1/S phases, S phase, G1/S transition, cell cycle checkpoints, M phase, generic transcription pathway, and metabolism are all heavily connected with common DEGs, according to Reactome’s pathways. The most prevalent DEGs connected with WikiPathways route include retinoblastoma gene in cancer, G1 to S cell cycle control, cell cycle, DNA IR-damage and cellular response via ATR, miRNA regulation of DNA damage response, vitamin D receptor pathway, and integrated breast cancer pathway.
Using common DEGs, a PPI network had been created to understand the biological characteristics in-depth and explore disease biomarkers. Depending on the topological measure (degree), 10 hub genes have been traced from the PPI network, which might be a therapeutic target or biomarker. The top 10 hub genes including CDK1, CCNB1, CDC20, TOP2A, MAD2L1, NDC80, AURKA, ASPM, NCAPG, and BIRC5 showed high degree value. In CC, cyclin-dependent kinase 1 (CDK1) has been observed before. CDK1 is a highly preserved protein that works as a serine kinase/threonine. With over 70 regulatory objectives, it plays a key role in controlling the cell cycle. CDK1 phosphorylates directly a number of target substrates for controlling the transcription and progression of cells in response to different stimuli [78]. Studies have demonstrated that CDKs and their modulators are aberrantly activated in several malignancies. CDK dysregulation induces the growth of aberrant cells and genomic instability [79]. Indeed, all human malignancies are affected by the D-cyclin-cdk4/6-INK4-Rb pathway [80]. Research from in vitro and in vivo shows that a variety of malignancies including cervical, colon, and breast cancer have substantial anticancer effects using CDK inhibitors [81, 82].
The transcriptional and posttranscriptional regulators of the hub DEGs were discovered using the miRNA-TF co-regulatory network. TFs govern transcription ratios, whereas miRNAs play a significant role in gene posttranscriptional regulation and RNA silence. The role of transcription factors (TFs) and microRNAs (miRNAs) in the progression of disease is crucial. Thus, the connections between the common DEGs, TFs, and miRNAs are shown in our research. The network builds on 73 TF candidates, 8 hub nodes, and 31 miRNA candidates. From the miRNA candidate, 4 candidates (hsa-mir-137, hsa-mir-92a, hsa-mir-24, and hsa-mir-542-3p) showed significant association with various types of cancer. miR-137 is integrated into CpG island (a high-frequency genomic area that contains CpG dinucleotide) and has been found in several types of cancer, such as colorectal, gastric, breast-and-squamous cells, and head and neck to have often been silenced by the promoter of hypermethylation [83–85]. MiR-137 is inhibited epigenetically in colorectal adenomatous cells in the same way as it is suppressed in colorectal cancer tissue, showing that miRNA methylation occurs early in colorectal cancer [86]. There are various studies which showed that hsa-mir-92a miRNA significantly connected with CC [87], colorectal cancer [88], small cell lung cancer [89], breast cancer [90], etc. According to some previous studies, hsa-mir-24 miRNA might play a significant role in lung cancer [91], breast cancer [92], prostate cancer [93], and colorectal cancer [94].
Gene-disease association network was analyzed to reveal the gene connectivity with the disease. The outcomes of this analysis showed liver carcinoma, lung carcinoma, adenocarcinoma, non-small cell lung carcinoma, renal cell carcinoma, colorectal neoplasms, prostatic neoplasms, neuroblastoma, kidney neoplasms, adrenocortical carcinoma, etc. are significantly connected with the hub DEGs. In individuals with CC, the history of radiation was an independent risk factor for second primary lung cancer [95]. A study indicated that 4.33% of CC patients develop lung metastasis [96], which is consistent with prior research [97, 98].
Another biggest exploration of this study is drug compounds finding for cancer. This analysis showed many compounds are connected with cancer. From them, LUCANTHONE CTD 00006227 and paclitaxel CTD 00007144 targets are significantly associated with CC. “LUCANTHONE” is the new target for CC. In previous, some studies have announced “LUCANTHONE” reduces cancer cell survival regardless of p53 status; autophagy suppression may be a more important contribution to the lucanthone mode of action that impacts DNA repair. Another drug compound target “paclitaxel” was studied as an anticancer medication in some previous studies [58–60]. The authors in these studies wrote, “paclitaxel” may play a significant role in CC as well as ovarian cancer, head and neck cancer, esophageal cancer, breast cancer, and lung cancer. Paclitaxel has a key role in the care of advanced/metastatic illness in cervical cancer, according to the European Society for Medical Oncology (ESMO) clinical practice guideline issued in 2017 [59]. Paclitaxel and cisplatin in combination with bevacizumab are regarded as the optimal first-line regimens for metastatic or recurrent cervical cancer due to the appropriate balance of effectiveness and safety characteristics. Paclitaxel with carboplatin may be an alternate choice for individuals who are not candidates for cisplatin, particularly those with impaired renal function [99]. Paclitaxel has been the first microtubule-stabilizing agent identified and considered as a significant part of the standard chemotherapy regimens for treating cervical cancer. In particular, the combination of paclitaxel with platinum-derived drugs has represented and represents today the cornerstone of advanced cervical cancer therapy, in particular for women with recurrent and persistent disease [59].
Conclusion
In this study, three datasets of CC were analyzed and compared to identify the important DEGs. A total of 315 common DEGs have collected for further analysis. Using common DEGs were used the functional analysis to figure out important GO terms and pathways using KEGG, Reactome, and WikiPathways database. Also, we developed a PPI network and identified significant hub DEGs including CDK1, CCNB1, CDC20, TOP2A, MAD2L1, NDC80, AURKA, ASPM, NCAPG, and BIRC5 that are the main therapeutic targets for the CC. And gene disease association and TF-miRNA co-regulatory network also demonstrated to identify the drag target miRNA. The analysis of the drug compounds showed LUCANTHONE CTD 00006227 and paclitaxel CTD 00007144 are the most associated with the hub DEGs. This network-based study may play a significant role in future treatment or medicine development for the patient with CC.
Acknowledgements
The authors would like to thank and extend their appreciation and gratitude to Al-Mustaqbal University College in Iraq for funding this project.
Authors’ contributions
MTH, MRI, and MRI carried out the experimental work and provided the first draft of the manuscript. KA, FMB, SA, and BRA supervised the experimental work and provided manuscript writing assistance. KA, MTH, and SA helped to revise the manuscript properly. KA, FMB, and MAM designed and supervised the work. The authors read and approved the final manuscript.
Funding
This work was supported by Al-Mustaqbal University College and in part by funding from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Md. Tanvir Hasan, Email: mdtanvir.hasan@ugent.be.
Md. Rakibul Islam, Email: rakibul35-116@diu.edu.bd.
Md. Rezwan Islam, Email: rezwan35-235@diu.edu.bd.
Baraa Riyadh Altahan, Email: Baraa.riyadh@mustaqbal-College.edu.iq.
Kawsar Ahmed, Email: kawsar.ict@mbstu.ac.bd, Email: k.ahmed.bd@ieee.org, Email: kawsarit08050@gmail.com, Email: k.ahmed@ussk.ca.
Francis M. Bui, Email: francis.bui@usask.ca
Sami Azam, Email: sami.azam@cdu.edu.au.
Mohammad Ali Moni, Email: m.moni@uq.edu.au.
References
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hricak H, Ward ZJ, Atun R, Abdel-Wahab M, Muellner A, Scott AM. Increasing access to imaging for addressing the global cancer epidemic. Radiology. 2021;301(3):543–546. doi: 10.1148/radiol.2021211351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang M, Zhong J, Zhang W, Zhou C, Wang X, Zou W, Xiaodan W, Zhang M. Psychometric properties of a simplified Chinese version of the cancer predisposition perception scale. Asia Pac J Oncol Nurs. 2022;9(3):179–184. doi: 10.1016/j.apjon.2021.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagai H, Kim YH. Cancer prevention from the perspective of global cancer burden patterns. J Thorac Dis. 2017;9(3):448–451. doi: 10.21037/jtd.2017.02.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain AM, Lafta RK. Cancer Trends in Iraq 2000-2016. Oman Med J. 2021;36(1):e219. doi: 10.5001/omj.2021.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carioli G, Bertuccio P, Malvezzi M, Rodriguez T, Levi F, Boffetta P, La Vecchia C, Negri E. Cancer mortality predictions for 2019 in Latin America. Int J Cancer. 2020;147(3):619–632. doi: 10.1002/ijc.32749. [DOI] [PubMed] [Google Scholar]
- 7.Ourlad AG, Tantengco YN, Yoshimura M, Nishiumi F, Llamas-Clark EF, Yanagihara I. Co-infection of human papillomavirus and other sexually transmitted bacteria in cervical cancer patients in the Philippines. Gynecol Oncol Rep. 2022;40:100943. doi: 10.1016/j.gore.2022.100943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadri Nahand J, Moghoofei M, Salmaninejad A, Bahmanpour Z, Karimzadeh M, Nasiri M, Mirzaei HR, Pourhanifeh MH, Bokharaei-Salim F, Mirzaei H, Hamblin MR. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer. 2020;146:305–320. doi: 10.1002/ijc.32688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dillman RK, Oldham RO, editors. Principles of cancer biotherapy. 5. Dordrecht: Springer; 2009. p. 149. [Google Scholar]
- 10.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ. International Agency for Research on Cancer Multicenter CC Study Group. Epidemiologic classification of human papillomavirus types associated with CC. N Engl J Med. 2003;348(6):518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 11.Srivastava P, Mangal M, Agarwal SM. Understanding the transcriptional regulation of cervix cancer using microarray gene expression data and promoter sequence analysis of a curated gene set. Gene. 2014;535(2):233–238. doi: 10.1016/j.gene.2013.11.028. [DOI] [PubMed] [Google Scholar]
- 12.Xue H, Sun Z, Wu W, Du D, Liao S. Identification of hub genes as potential prognostic biomarkers in cervical cancer using comprehensive bioinformatics analysis and validation studies. Cancer Manag Res. 2021;13:117–131. doi: 10.2147/CMAR.S282989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindumathi V, Kranthi T, Rao SB, Manimaran P. The prediction of candidate genes for cervix related cancer through gene ontology and graph theoretical approach. Mol Biosyst. 2014;10(6):1450–1460. doi: 10.1039/c4mb00004h. [DOI] [PubMed] [Google Scholar]
- 14.Jalan S, Kanhaiya K, Rai A, Bandapalli OR, Yadav A. Network topologies decoding cervical cancer. PLoS One. 2015;10(8):e0135183. doi: 10.1371/journal.pone.0135183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Li Z, Gao A, Wen Q, Sun Y. The prognostic landscape of tumor-infiltrating immune cells in cervical cancer. Biomed Pharmacother. 2019;120:109444. doi: 10.1016/j.biopha.2019.109444. [DOI] [PubMed] [Google Scholar]
- 16.Pahne-Zeppenfeld J, Schröer N, Walch-Rückheim B, Oldak M, Gorter A, Hegde S, Smola S. Cervical cancer cell-derived interleukin-6 impairs CCR7-dependent migration of MMP-9-expressing dendritic cells. Int J Cancer. 2014;134:2061–2073. doi: 10.1002/ijc.28549. [DOI] [PubMed] [Google Scholar]
- 17.Kori M, Yalcin AK. Potential biomarkers and therapeutic targets in cervical cancer: insights from the meta-analysis of transcriptomics data within network biomedicine perspective. PLoS One. 2018;13(7):e0200717. doi: 10.1371/journal.pone.0200717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clough E, Barrett T (2016) The Gene Expression Omnibus Database. Methods Mol Biol 1418:93-110. 10.1007/978-1-4939-3578-9_5 [DOI] [PMC free article] [PubMed]
- 19.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau W-C, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33(Issue suppl_1):D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilhite SE, Barrett T (2012) Strategies to explore functional genomics data sets in NCBI's GEO database. Methods Mol Biol 802:41–53. 10.1007/978-1-61779-400-1_3 [DOI] [PMC free article] [PubMed]
- 21.Scotto L, Narayan G, Nandula SV, Arias-Pulido H, et al. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in CC: potential role in progression. Genes Chromosomes Cancer. 2008;47(9):755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 22.Den Boon JA, Pyeon D, Wang SS, Horswill M, et al. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112(25):E3255–E3264. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhai Y, Kuick R, Nan B, Ota I, et al. Gene expression analysis of preinvasive and invasive cervical squamous cell carcinomas identifies HOXC10 as a key mediator of invasion. Cancer Res. 2007;67(21):10163–10172. doi: 10.1158/0008-5472.CAN-07-2056. [DOI] [PubMed] [Google Scholar]
- 24.Blount JR, Meyer DN, et al. Una` nchored ubiquitin chains do not lead to marked alterations in gene expression in Drosophila melanogaster. Biol Open. 2019;8:bio043372. doi: 10.1242/bio.043372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Heather Butler J, Cherry M, Davis AP, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Avi Ma’ayan. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14(1):128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14:128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed]
- 29.Tanvir Hasan M, Hassan M, Kawsar Ahmed M, Islam R, Islam K, Bhuyian T, Uddin MS, Paul BK. Network based study to explore genetic linkage between diabetes mellitus and myocardial ischemia: bioinformatics approach. Gene Rep. 2020;21:100809. doi: 10.1016/j.genrep.2020.100809. [DOI] [Google Scholar]
- 30.Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A et al (2016) The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res:gkw937. 10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bader GD, Hogue CWV. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf. 2003;4(1):2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chin C-H, Chen S-H, Hsin-Hung W, Ho C-W, Ko M-T, Lin C-Y. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(S4):S11. doi: 10.1186/1752-0509-8-S4-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoo M, Shin J, Kim J, Ryall KA, Lee K, Lee S, Jeon M, Kang J, Tan AC. DSigDB: drug signatures database for gene set analysis. Bioinformatics. 2015;31(18):3069–3071. doi: 10.1093/bioinformatics/btv313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dawn Teare M, Barrett JH. Genetic linkage studies. Lancet. 2005;366(9490):1036–1044. doi: 10.1016/S0140-6736(05)67382-5. [DOI] [PubMed] [Google Scholar]
- 36.Frayling TM. Genome-wide association studies provide new insights into type 2 diabetes aetiology. Nat Rev Genet. 2007;8(9):657–662. doi: 10.1038/nrg2178. [DOI] [PubMed] [Google Scholar]
- 37.Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9(7):554–566. doi: 10.1038/nrg2364. [DOI] [PubMed] [Google Scholar]
- 38.Piro RM, Di Cunto F. Computational approaches to disease-gene prediction: rationale, classification and successes. FEBS J. 2012;279(5):678–696. doi: 10.1111/j.1742-4658.2012.08471.x. [DOI] [PubMed] [Google Scholar]
- 39.Tranchevent LC, Capdevila FB, Nitsch D, De Moor B, De Causmaecker P, Moreau Y. A guide to web tools to prioritize candidate genes. Brief Bioinform. 2011;12(1):22–32. doi: 10.1093/bib/bbq007. [DOI] [PubMed] [Google Scholar]
- 40.Oti M, Ballouz S, Wouters MA. Web tools for the prioritization of candidate disease genes. Methods Mol Biol. 2011;760:189–206. doi: 10.1007/978-1-61779-176-5_12. [DOI] [PubMed] [Google Scholar]
- 41.Opap K, Mulder N. Recent advances in predicting gene-disease associations. F1000Res. 2017;6:578. doi: 10.12688/f1000research.10788.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohamed RH, Abu-Shahba N, Mahmoud M, Abdelfattah AMH, Zakaria W, ElHefnawi M. Co-regulatory network of oncosuppressor miRNAs and transcription factors for pathology of human hepatic cancer stem cells (HCSC) Sci Rep. 2019;9(1):5564. doi: 10.1038/s41598-019-41978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu ZP, Wu C, Miao H, Wu H. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford) 2015;2015:bav095. doi: 10.1093/database/bav095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schober P, Vetter TR. Survival analysis and interpretation of time-to-event data: the tortoise and the hare. Anesth Analg. 2018;127(3):792–798. doi: 10.1213/ane.0000000000003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George B, Seals S, Aban I. Survival analysis and regression models. J Nucl Cardiol. 2014;21(4):686–694. doi: 10.1007/s12350-014-9908-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.In J, Lee DK. Survival analysis: part II - applied clinical data analysis. Korean J Anesthesiol. 2019;72(5):441–457. doi: 10.4097/kja.19183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koletsi D, Pandis N. Survival analysis, part 3: cox regression. Am J Orthod Dentofacial Orthop. 2017;152(5):722–723. doi: 10.1016/j.ajodo.2017.07.009. [DOI] [PubMed] [Google Scholar]
- 48.Luo M, Kelley MR. Inhibition of the human apurinic/apyrimidinic endonuclease (APE1) repair activity and sensitization of breast cancer cells to DNA alkylating agents with lucanthone. Anticancer Res. 2004;24(4):2127–2134. [PubMed] [Google Scholar]
- 49.Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29(3):451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carew JS, Medina EC, Esquivel JA, 2nd, Mahalingam D, Swords R, Kelly K, Zhang H, Huang P, Mita AC, Mita MM, Giles FJ, Nawrocki ST. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010;14(10):2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, Houghton JA, Huang P, Giles FJ, Cleveland JL. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu L, Chen L. Progress in research on paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 2019;24:40. doi: 10.1186/s11658-019-0164-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vassileva V, Allen CJ, Piquette-Miller M. Effects of sustained and intermittent paclitaxel therapy on tumor repopulation in ovarian cancer. Mol Cancer Ther. 2008;7(3):630–637. doi: 10.1158/1535-7163.MCT-07-2117. [DOI] [PubMed] [Google Scholar]
- 54.Giuseppe G, Arun R, Arlene B, Kelly RJ, Szabo E, Ariel LC, et al. Phase II study of belinostat in patients with recurrent or refractory advanced Thymic epithelial tumors. J Clin Oncol. 2011;29(15):2052–2059. doi: 10.1200/JCO.2010.32.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hazem G, Cynthia L, Eman B, Khaldoon AR, Asma T, Monther AA, Hendrayani SF, Pulicat M, Ayodele A, Taher AT, Abdelilah A, Said D. Research article doxorubicin downregulates cell surface B7-H1expression and upregulates its nuclear expression in breast cancer cells: role of B7-H1 as an anti-apoptotic molecule. Breast Cancer Res. 2010;12:4. doi: 10.1186/bcr2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tan JL, Chan ST, Lo CY, Deane JA, McDonald CA, Bernard CC, Wallace EM, Lim R. Amnion cell-mediated immune modulation following bleomycin challenge: controlling the regulatory T cell response. Stem Cell Res Ther. 2015;6(1):8. doi: 10.1186/scrt542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koreth J, Stevenson KE, Kim HT, McDonough SM, Bindra B, Armand P, Ho VT, Cutler C, Blazar BR, Antin JH, Soiffer RJ, Ritz J, Alyea EP., 3rd Bortezomib-based graft-versus-host disease prophylaxis in HLA-mismatched unrelated donor transplantation. J Clin Oncol. 2012;30(26):3202–3208. doi: 10.1200/JCO.2012.42.0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moore DH, Blessing JA, McQuellon RP, Thaler HT, Cella D, Benda J, Miller DS, Olt G, King S, Boggess JF, Rocereto TF. Phase III study of cisplatin with or without paclitaxel in stage IVB, recurrent, or persistent squamous cell carcinoma of the cervix: a gynecologic oncology group study. J Clin Oncol. 2004;22(15):3113–3119. doi: 10.1200/JCO.2004.04.170. [DOI] [PubMed] [Google Scholar]
- 59.Della Corte L, Barra F, Foreste V, Giampaolino P, Evangelisti G, Ferrero S, Bifulco G. Advances in paclitaxel combinations for treating CC. Expert Opin Pharmacother. 2020;21(6):663–677. doi: 10.1080/14656566.2020.1724284. [DOI] [PubMed] [Google Scholar]
- 60.Heeren AM, van Luijk IF, Lakeman J, Pocorni N, Kole J, de Menezes RX, Kenter GG, Bosse T, de Kroon CD, Jordanova ES. Neoadjuvant cisplatin and paclitaxel modulate tumor-infiltrating T cells in patients with CC. Cancer Immunol Immunother. 2019;68(11):1759–1767. doi: 10.1007/s00262-019-02412-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abu-Rustum NR, Yashar CM, Bean S, Bradley K, Campos SM, Chon HS, Chu C, Cohn D, Crispens MA, Damast S, Fisher CM, Frederick P, Gaffney DK, Giuntoli R, Han E, Huh WK, Lurain Iii JR, Mariani A, Mutch D, Nagel C, Nekhlyudov L, Fader AN, Remmenga SW, Reynolds RK, Sisodia R, Tillmanns T, Ueda S, Urban R, Wyse E, McMillian NR, Motter AD. NCCN Guidelines Insights: Cervical Cancer, Version 1.2020. J Natl Compr Canc Netw. 2020;18(6):660–666. doi: 10.6004/jnccn.2020.0027. [DOI] [PubMed] [Google Scholar]
- 62.Zighelboim I, Wright JD, Gao F, Case AS, Massad LS, Mutch DG, Powell MA, Thaker PH, Eisenhauer EL, Cohn DE, Valea FA, Alvarez Secord A, Lippmann LT, Dehdashti F, Rader JS. Multicenter phase II trial of topotecan, cisplatin and bevacizumab for recurrent or persistent cervical cancer. Gynecol Oncol. 2013;130(1):64–68. doi: 10.1016/j.ygyno.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin Y, Sibanda VL, Zhang HM, Hu H, Liu H, Guo AY. MiRNA and TF co-regulatory network analysis for the pathology and recurrence of myocardial infarction. Sci Rep. 2015;5:9653. doi: 10.1038/srep09653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chi Y, Xue J, Huang S, Xiu B, Su Y, Wang W, Guo R, Wang L, Li L, Shao Z, Jin W, Wu Z, Wu J. CapG promotes resistance to paclitaxel in breast cancer through transactivation of PIK3R1/P50. Theranostics. 2019;9(23):6840–6855. doi: 10.7150/thno.36338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bahrami S, Gheysarzadeh A, Sotoudeh M, Bandehpour M, Khabazian R, Zali H, Hedayati M, Basiri A, Kazemi B. The association between gelsolin-like actin-capping protein (CapG) overexpression and bladder cancer prognosis. Urol J. 2020;18(2):186–193. doi: 10.22037/uj.v0i0.5664. [DOI] [PubMed] [Google Scholar]
- 66.Jiang S, Yang Y, Zhang Y, Ye Q, Song J, Zheng M, Li X. Overexpression of CAPG is associated with poor prognosis and immunosuppressive cell infiltration in ovarian cancer. Dis Markers. 2022;2022:9719671. doi: 10.1155/2022/9719671. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Ma X, Zhang Q, Du J, Tang J, Tan B. Integrated analysis of ceRNA regulatory network associated with tumor stage in cervical cancer. Front Genet. 2021;12:618753. doi: 10.3389/fgene.2021.618753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xue JM, Liu Y, Wan LH, Zhu YX. Comprehensive analysis of differential gene expression to identify common gene signatures in multiple cancers. Med Sci Monit. 2020;26:e919953. doi: 10.12659/MSM.919953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D, Liu Y, Cheng S, Liu G. Identification of novel genes and associated drugs in cervical cancer by bioinformatics methods. Med Sci Monit. 2022;28:e934799. doi: 10.12659/MSM.934799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rahman MR, Islam T, Zaman T, Shahjaman M, Karim MR, Huq F, Quinn JMW, Holsinger RMD, Gov E, Moni MA. Identification of molecular signatures and pathways to identify novel therapeutic targets in Alzheimer's disease: Insights from a systems biomedicine perspective. Genomics. 2020;112(2):1290–1299. doi: 10.1016/j.ygeno.2019.07.018. [DOI] [PubMed] [Google Scholar]
- 71.Sainz B, Jr, Mossel EC, Peters CJ, Garry RF. Interferon-beta and interferon-gamma synergistically inhibit the replication of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) Virology. 2004;329(1):11–17. doi: 10.1016/j.virol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bergmann CC, Parra B, Hinton DR, Ramakrishna C, Dowdell KC, Stohlman SA. Perforin and gamma interferon-mediated control of coronavirus central nervous system infection by CD8 T cells in the absence of CD4 T cells. J Virol. 2004;78(4):1739–1750. doi: 10.1128/jvi.78.4.1739-1750.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cammack R, Atwood T, Campbell P, Parish H, Smith A, Vella F, Stirling J. Oxford dictionary of biochemistry and molecular biology. Oxford University Press; 2008. [Google Scholar]
- 74.Desvergne B, Michalik L, Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86(2):465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- 75.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103(5):1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tessitore A, Cicciarelli G, Del Vecchio F, Gaggiano A, Verzella D, Fischietti M, Vecchiotti D, Capece D, Zazzeroni F, Alesse E. MicroRNAs in the DNA damage/repair network and cancer. Int J Genomics. 2014;2014:820248. doi: 10.1155/2014/820248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hasan Mahmud SM, Al-Mustanjid M, Farzana Akter M, Rahman S, Kawsar Ahmed M, Rahman H, Chen W, Moni MA (2021) Bioinformatics and system biology approach to identify the influences of SARS-CoV-2 infections to idiopathic pulmonary fibrosis and chronic obstructive pulmonary disease patients. Brief Bioinform:bbab115. 10.1093/bib/bbab115 [DOI] [PMC free article] [PubMed]
- 78.Chatr-Aryamontri A, Breitkreutz BJ, Heinicke S, Boucher L, Winter A, Stark C, Nixon J, Ramage L, Kolas N, O'Donnell L, Reguly T, Breitkreutz A, Sellam A, Chen D, Chang C, Rust J, Livstone M, Oughtred R, Dolinski K, Tyers M. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41(Database issue):D816–D823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petignat P, Roy M. Diagnosis and management of CC. BMJ. 2007;335(7623):765–768. doi: 10.1136/bmj.39337.615197.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26(19):2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sheu BC, Lien HC, Ho HN, Lin HH, Chow SN, Huang SC, Hsu SM. Increased expression and activation of gelatinolytic matrix metalloproteinases is associated with the progression and recurrence of human CC. Cancer Res. 2003;63(19):6537–6542. [PubMed] [Google Scholar]
- 82.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 83.Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 84.Ando T, Yoshida T, Enomoto S, Asada K, Tatematsu M, Ichinose M, Sugiyama T, Ushijima T. DNA methylation of microRNA genes in gastric mucosae of gastric cancer patients: its possible involvement in the formation of epigenetic field defect. Int J Cancer. 2009;124(10):2367–2374. doi: 10.1002/ijc.24219. [DOI] [PubMed] [Google Scholar]
- 85.Vrba L, Muñoz-Rodríguez JL, Stampfer MR, Futscher BW. miRNA gene promoters are frequent targets of aberrant DNA methylation in human breast cancer. PLoS One. 2013;8(1):e54398. doi: 10.1371/journal.pone.0054398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, Goel A. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70(16):6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kong Q, Tang Z, Xiang F, et al. Diagnostic value of serum hsa-mir-92a in patients with CC. Clin Lab. 2017;63(2):335–340. doi: 10.7754/clin.lab.2016.160610. [DOI] [PubMed] [Google Scholar]
- 88.Fangfang F, Jiang W, Zhou L, Chen Z. Circulating exosomal miR-17-5p and miR-92a-3p predict pathologic stage and grade of colorectal cancer. Translat Oncol. 2018;11(2):221–232. doi: 10.1016/j.tranon.2017.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu Y, et al. Plasma miR-92a-2 as a biomarker for small cell lung cancer. 2017. pp. 319–327. [DOI] [PubMed] [Google Scholar]
- 90.Cun J, Yang Q. Bioinformatics-based interaction analysis of miR-92a-3p and key genes in tamoxifen-resistant breast cancer cells. Biomed Pharmacother. 2018;107:117–128. doi: 10.1016/j.biopha.2018.07.158. [DOI] [PubMed] [Google Scholar]
- 91.Pan Y, Wang H, Ma D, Ji Z, Luo L, Cao F, Huang F, Liu Y, Dong Y, Chen Y. miR-24 may be a negative regulator of menin in lung cancer. Oncol Rep. 2018;39(5):2342–2350. doi: 10.3892/or.2018.6327. [DOI] [PubMed] [Google Scholar]
- 92.Han X, Li Q, Liu C, Wang C, Li Y. Overexpression miR-24-3p repressed Bim expression to confer tamoxifen resistance in breast cancer. J Cell Biochem. 2019;120(8):12966–12976. doi: 10.1002/jcb.28568. [DOI] [PubMed] [Google Scholar]
- 93.Lynch SM, McKenna MM, Walsh CP, McKenna DJ. miR-24 regulates CDKN1B/p27 expression in prostate cancer. Prostate. 2016;76(7):637–648. doi: 10.1002/pros.23156. [DOI] [PubMed] [Google Scholar]
- 94.Hao JP, Ma A. The ratio of miR-21/miR-24 as a promising diagnostic and poor prognosis biomarker in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(24):8649–8656. doi: 10.26355/eurrev_201812_16629. [DOI] [PubMed] [Google Scholar]
- 95.Qian C, Liu H, Feng Y, Meng S, Wang D, Nie M, Xu M. Clinical characteristics and risk of second primary lung cancer after CC: a population-based study. PLoS One. 2020;15(8):e0231807. doi: 10.1371/journal.pone.0231807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen X, Chen L, Zhu H, Tao J. Risk factors and prognostic predictors for CC patients with lung metastasis. J Cancer. 2020;11(20):5880–5889. doi: 10.7150/jca.46258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ki EY, Lee KH, Park JS, Hur SY. A clinicopathological review of pulmonary metastasis from uterine CC. Cancer Res Treat. 2016;48(1):266–272. doi: 10.4143/crt.2014.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zheng A, Chen Y, Fang J, Zhang Y. Clinicopathologic characteristics and risk factors for lung metastasis after radical hysterectomy in early-stage CC. Zhonghua Fu Chan Ke Za Zhi. 2015;50(3):204–209. [PubMed] [Google Scholar]
- 99.Marth C, Landoni F, Mahner S, McCormack M, Gonzalez-Martin A, Colombo N, ESMO Guidelines Committee Cervical cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2017;28(suppl_4):iv72–iv83. doi: 10.1093/annonc/mdx220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.