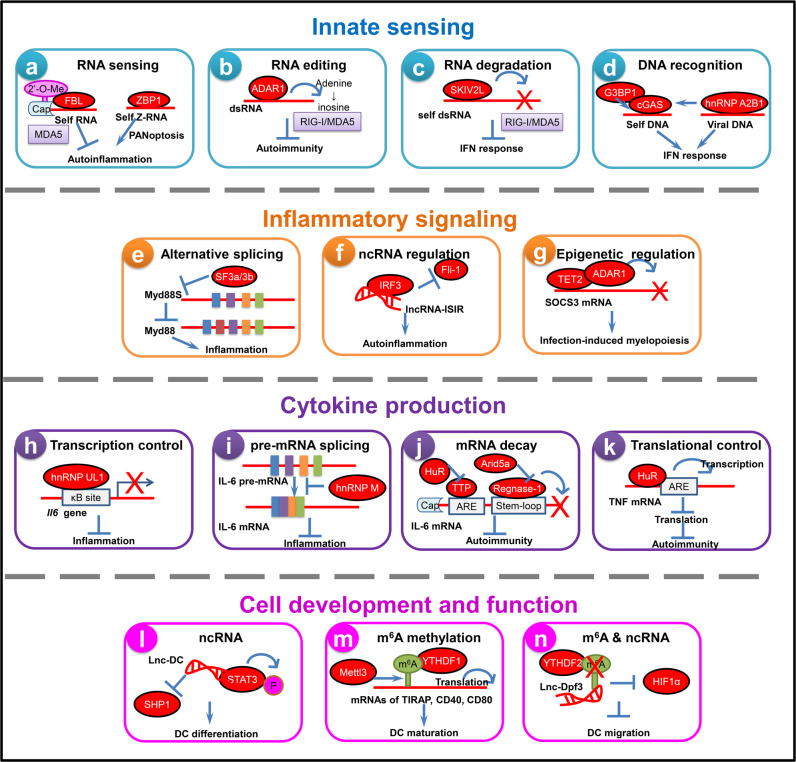
Fig. 2. Control of RNA metabolism and function by RBPs in innate immune response during autoimmunity and autoinflammation.
RBPs regulate the activation of PRR-triggered innate immunity via targeting various steps at transcriptional, post-transcriptional or translational levels. a FBL mediates 2′-O-methylation of RNA and prevents the innate recognition by MDA5 and IFN responses. ZBP1 recognizes self Z-RNA to promote pathologic inflammation. b ADAR1 mediates RNA A-to-I editing, thus allowing effective antiviral immunity while preventing pathogenic autoinflammation. c The SKIV2L subunit of the RNA exosome cleaves self RNA produced by the endonuclease IRE-1, and thereby inhibits RIG-I activation and type I IFN-dependent autoinflammation. d G3BP1 enhances DNA binding of cGAS and cGAS-dependent IFN production. hnRNP A2B1 recognizes viral DNA and enhances cGAS/STING-dependent IFN response. e The short isoform of MyD88 (MyD88s) inhibits TLR-triggered inflammation due to its failure to recruit IRAK-4. SF3a and SF3b mRNA splicing complexes reduce the MyD88s mRNA levels. f lncRNA-ISIR binds to IRF3 and impedes the inhibitory effect of Fli-1, thus enhancing IRF3 activation, IFN response and autoinflammation. g TET2 promotes the degradation of SOCS3 mRNA through ADAR1, facilitating cytokine-induced emergency myelopoiesis and mast cell expansion and activation during pathogen infection. h hnRNP UL1 inhibits NF-κB-mediated inflammation via competing with NF-κB to bind κB sites, while hnRNP UL1 expression decreases in RA patients. i hnRNP M inhibits pre-mRNA splicing and maturation of inflammatory transcripts such as IL-6 to negatively regulate inflammatory responses. j TTP and Regnase-1 destabilize mRNAs of proinflammatory cytokines such as IL-6 and TNF to control autoimmunity. k HuR downregulates mRNA translation to suppress aberrant inflammation and autoimmunity. l lnc-DC controls human DC differentiation and function via directly binding to STAT3 in the cytoplasm to prevent SHP1 binding and promote STAT3 phosphorylation. m Mettl3 mediates m6A methylation of transcripts of co-stimulatory molecules CD40, CD80 and TLR4 signaling adaptor TIRAP to enhance their translation in DCs, stimulating T cell activation and strengthening TLR4/NF-κB signaling-induced cytokine production. n CCR7 ligation upregulates expression of lnc-Dpf3 via relieving m6A-dependent degradation, consequently leading to inhibition of HIF1α-dependent glycolysis and DC migration. RBPs responsible for each step are shown in red ovals. FBL fibrillarin, ZBP1 Z-DNA-binding protein 1, MDA5 melanoma differentiation-associated gene 5, ADAR1 adenosine deaminase acting on RNA 1, RIG-I retinoic acid-inducible gene I, SKIV2L superkiller viralicidic activity 2-like, G3BP1 GTPase-activating protein SH3 domain-binding protein 1, cGAS cyclic GMP AMP synthase, hnRNA A2B1 heterogeneous nuclear ribonucleoprotein A2B1, MyD88 Myeloid differentiation primary response gene 88, SF3a/3b splicing factor 3a/3b, IRF3 interferon regulatory factor 3, Fli-1 flightless-1, TET2 ten-eleven translocation 2, hnRNP UL1 heterogeneous nuclear ribonucleoprotein UL1, hnRNP M heterogeneous nuclear ribonucleoprotein M, TTP tristetraprolin, HuR human antigen R, Arid5a AT-rich interactive domain-containing protein 5a, ARE adenine uridine (AU)-rich elements, DC dendritic cells, STAT3 signal transducer and activator of transcription 3, SHP1 Src-homology 2 (SH2) domain-containing phosphatase 1, Mettl3 methyltransferase like 3, YTHDF1/2 YTH domain-containing protein 1/2, HIF1α hypoxia-inducible factor-1 alpha.