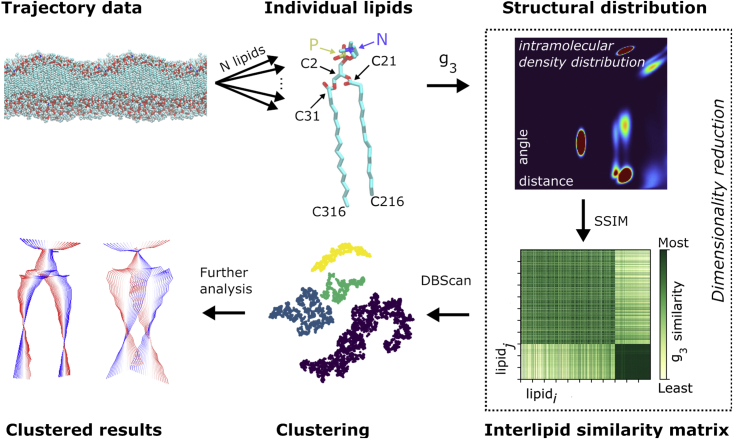
Figure 2.
Conformational clustering. First, each lipid (some of the important atoms are marked) is isolated from the trajectory and is calculated to quantify the intramolecular structure. The mean structural similarity index metric (SSIM), Eq. 2, of each lipid distribution is compared with all other lipids. Embedding with t-SNE (33) reduces the matrix of similarity values to a 2D form (from dimensions). These 2D data are clustered using DBScan (34) to find similar conformational groupings. This is mapped back onto the bilayer for further analysis. The similarity matrix shows how similar the distribution of row is with the distribution of row . The distinct difference in the final quarter of the similarity matrix is a result of passing over the phase transition and the large conformation difference this causes. To see this figure in color, go online.