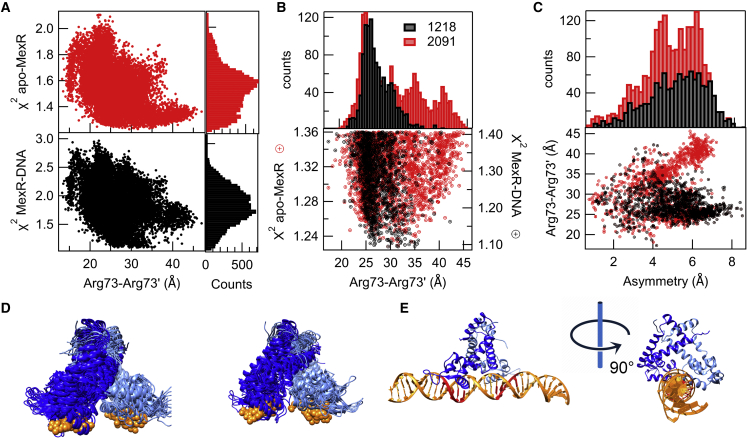
Figure 4.
SAS-based molecular modeling. (A) Distribution of MD-derived MexR models as a function of the Arg73-Arg73′ distance and to the SAXS data of the apo-protein (red) and the SANS data of dMexR-PII at 56% D2O (black). Model counts show the propensities of models with best fit to data as a function of to the corresponding data. (B) Magnification of best-fitting models (15% of total range), colored as in (A), with model counts as a function of the Arg73-Arg73′ distances; the ensembles comprise 2091 structures for apo-MexR and 1218 structures for MexR when bound to DNA. 63% (770 structures) of the structures in the MexR-DNA ensemble are also members of the apo-MexR ensemble. (C) Scatter plot of the asymmetry of the best 15% over the Arg73 distances. In red are the structures for the apo-MexR and in black the MexR in the presence of the DNA. From the scatter plot, it is evident that MexR in the presence of the DNA presents more asymmetry than in absence of the DNA. (D) Best-fitting ensembles representing apo-MexR and MexR when bound to DNA (MexR-DNA); 21/10 structures were chosen randomly from the 2091/1218 structures of the respective ensembles shown in (B)–(C). Monomeric units of the MexR are in blue/skyblue, and Arg73/Arg73′ sidechains are shown in orange spheres. (E) Best-fitting MexR-DNA complex to the combined SANS/SAXS data of MexR-DNA (overlaid with ab initio modeling in Fig. 4B). The positioning of the palindromes is highlighted in red. To see this figure in color, go online.