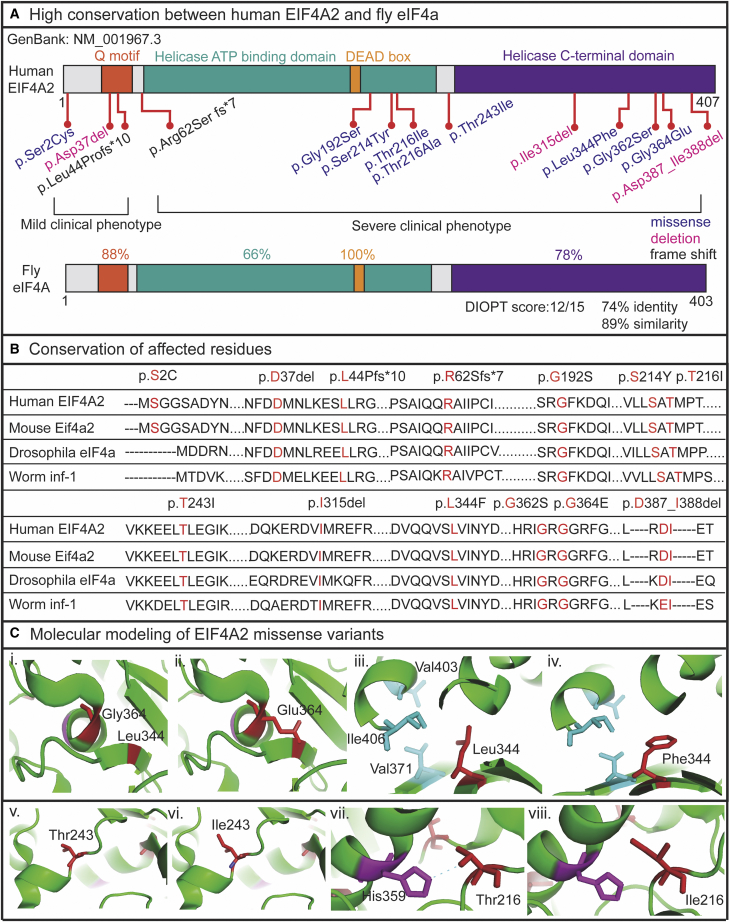
Figure 3.
The EIF4A2 variants are well conserved across different species
(A) EIF4A2 variants are positioned in the corresponding protein domains (missense variants are shown in purple, deletion variants are shown in pink, and the frameshift variants are shown in black). The fruit fly homolog, eIf4A shows 74% identity and 89% similarity with 88% matching in Q motif, 66% matching in helicase ATP-binding domain, 100% matching in DEAD box, and 78% matching in helicase C-terminal domain.
(B) Conservation analysis of affected residues in different species are shown.
(C) Molecular modeling of four of the missense variants are shown and the critical residues are colored in red. (i, iii, v, vii) shows the WT residues and (ii, iv, vi, viii) shows the variant residues.