A list of marine-derived antimalarial alkaloids showing their IC50 against various strains of Plasmodium sp., chemical structure and biogenic sourcea.
| Compound | Antiplasmodial activity (IC50 value) | Structure | Source | Marine class | Ref. |
|---|---|---|---|---|---|
| Manzamine A (1) | W2 = 0.015 μM D6 = 0.0082 μM |
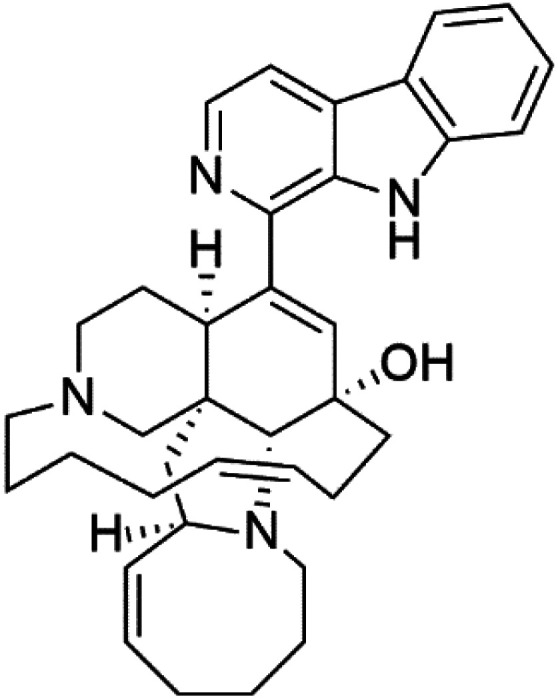
|
Okinawan Haliclona | Sponge | 27 |
| 8-Hydroxymanzamine (2) | W2 = 0.014 μM D6 = 0.010 μM |
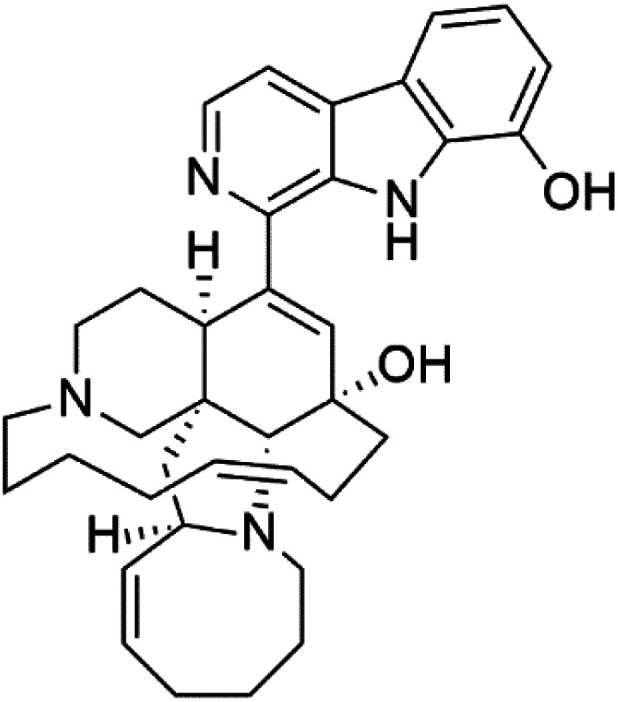
|
Sponge | 27 | |
| Manzamine F (3) | W2 = 2.93 μM 6 = 1.34 μM |
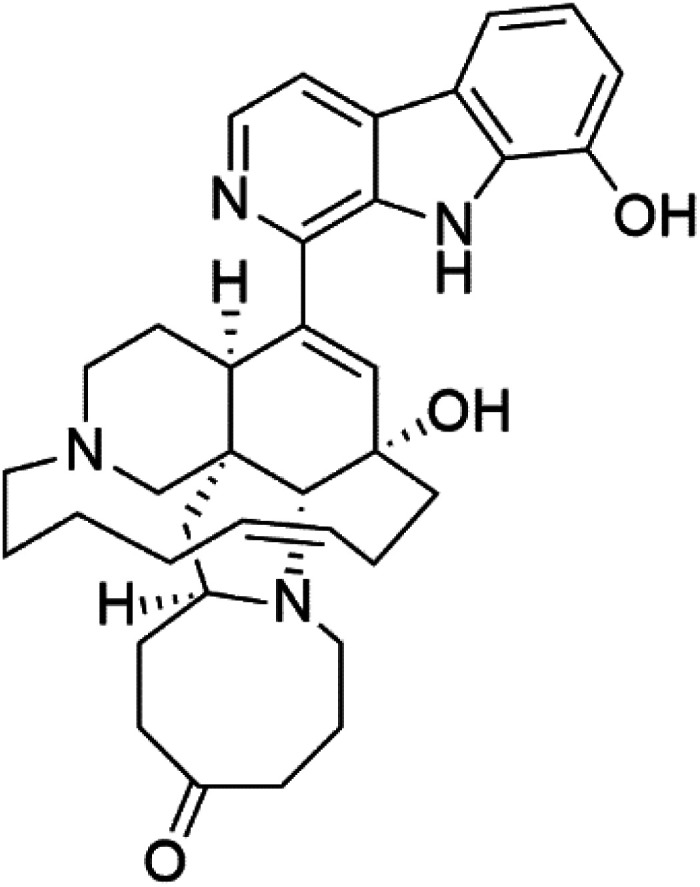
|
Sponge | 27 | |
| 6-Hydroxymanzamine (4) | W2 = 1.5 μM D6 = 1.36 μM |
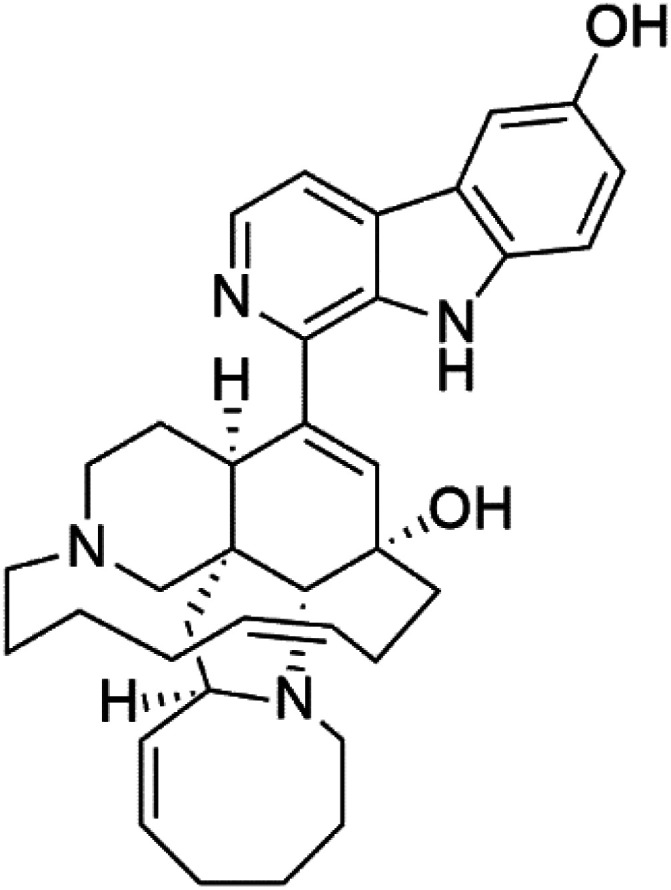
|
Sponge | 27 | |
| Neo-kauluamine (5) | D6 = 1.46 μM W2 = 2.41 μM |
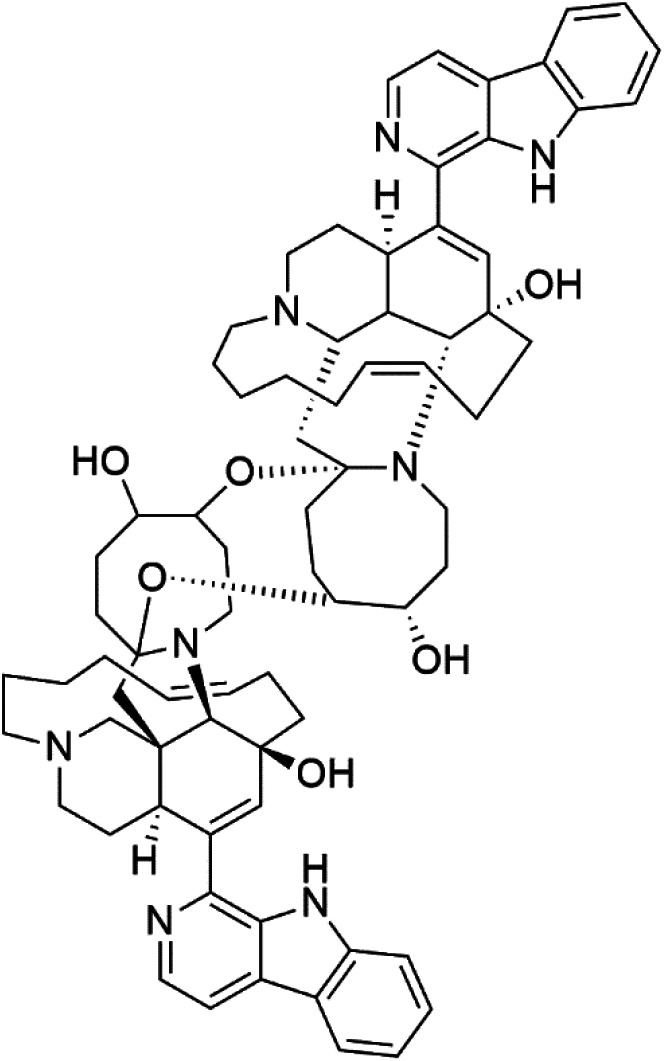
|
Indo-pacific sponge | Sponge | 28 |
| 12,34-Oxamanzamine A (6) | D6 = 8.97 μM |
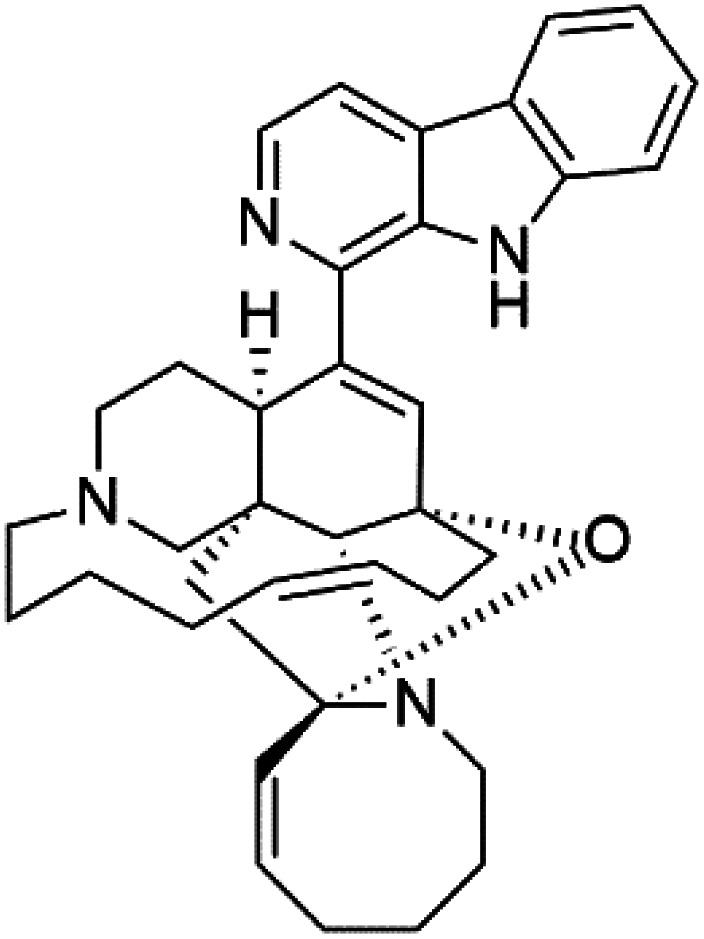
|
Sponge | 28 | |
| Zamamidine A (7) | 0.0008 to 0.016 μM |
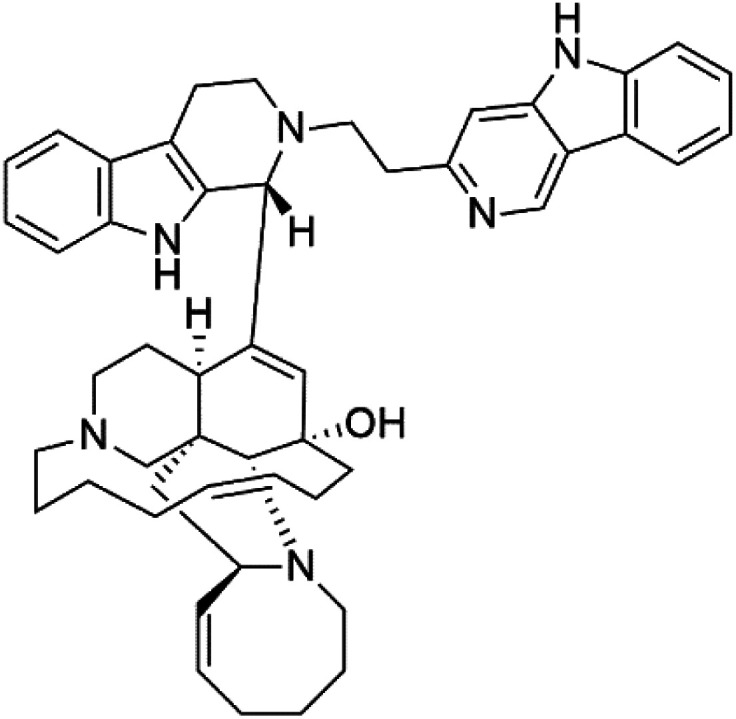
|
Amphimedon sp. | Sponge | 35 |
| Zamamidine B (8) |
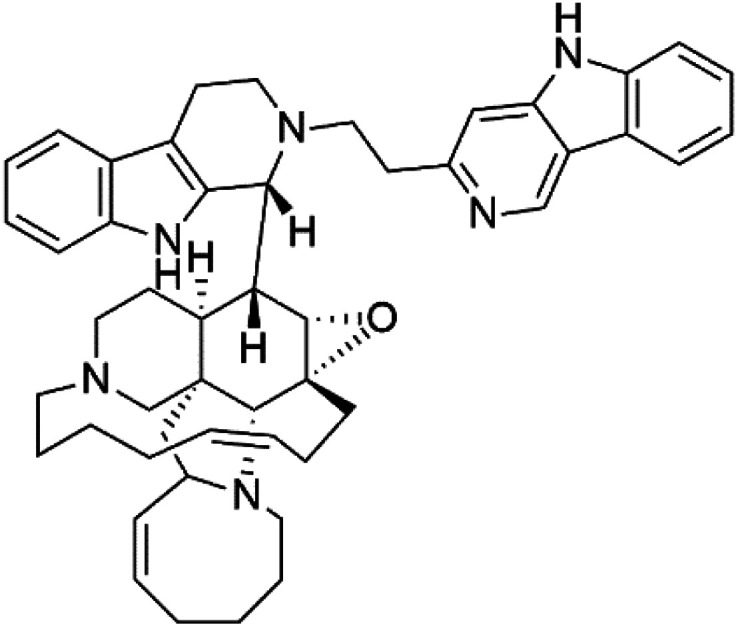
|
Sponge | |||
| Zamamidine C (9) |
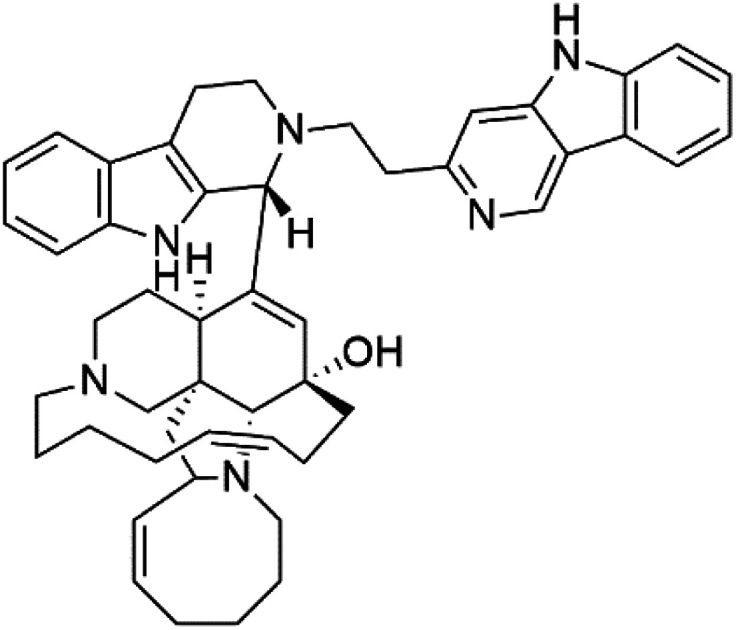
|
Sponge | |||
| Zamamidine D (10) |
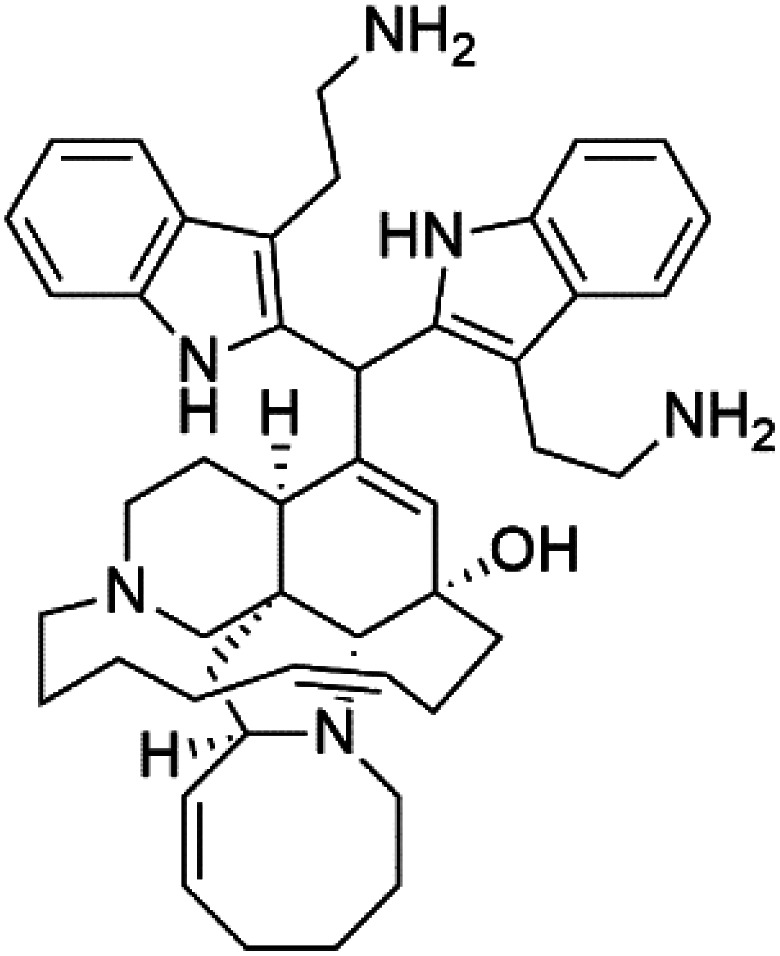
|
Sponge | |||
| Homofascaplysin (11) | K1 = 0.04 μM NF54 = 0.07 μM |
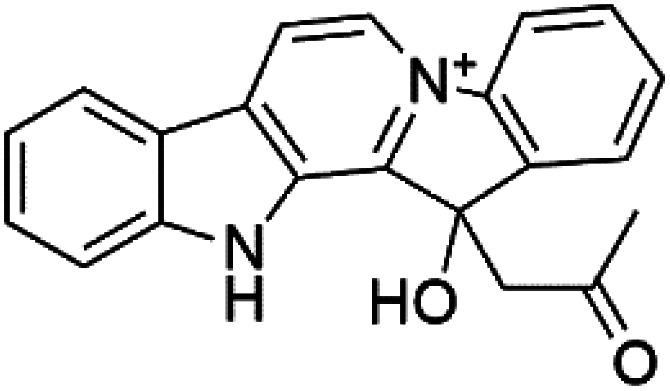
|
Hyrtios sponge | Sponge | 38 |
| Marinacarboline A (12) Marinacarboline B (13) Marinacarboline C (14) | 3D7 and Dd2 IC50 from 1.92 to 36.03 μM |
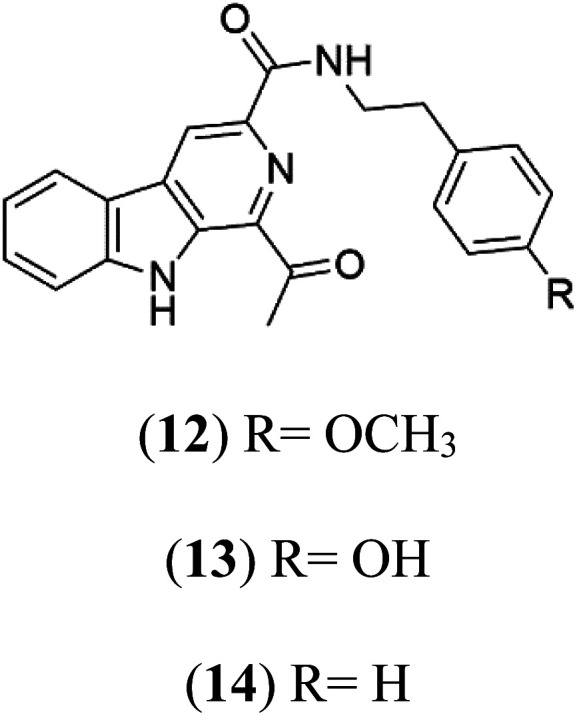
|
Marinactinospora thermotolerans | Actinomycete bacteria | 39 |
| Marinacarboline D (15) |
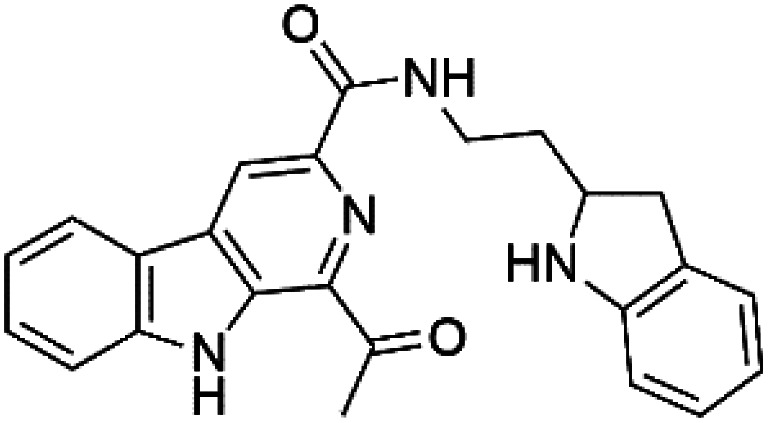
|
||||
| 13-N-Demethyl-methylpendolmycin (16) methylpendolmycin-14-O-α-glucoside (17) | 3D7 = 20.75 and 10.43 μM Dd2 = 18.67 and 5.03 μM |
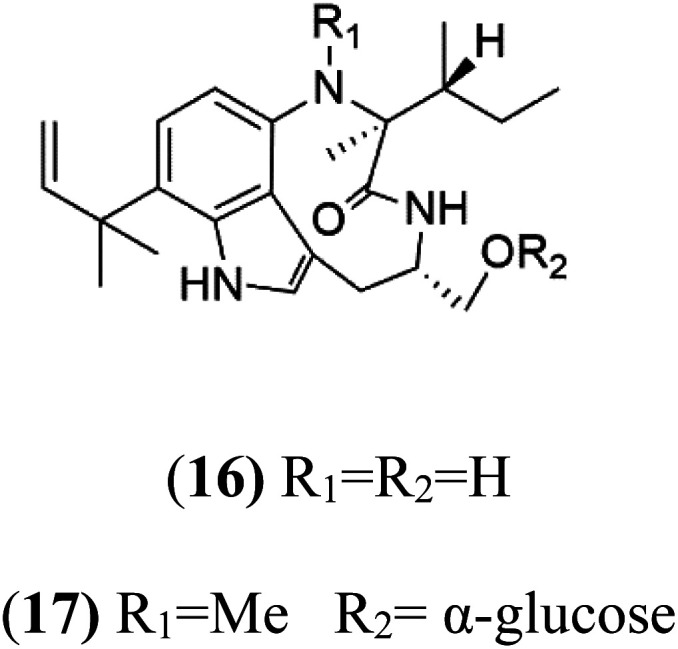
|
Marinactinospora thermotolerans | Actinomycete bacteria | 38 |
| Crambescidin 800 (18) | 3D7 = 0.16 μM FCR3 = 0.24 μM |
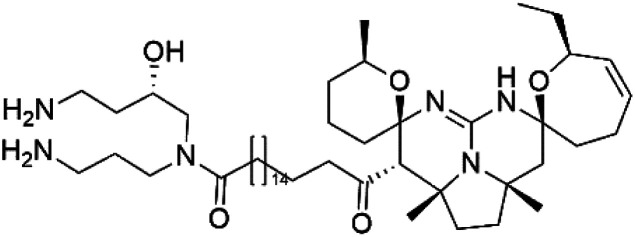
|
Monanchora unguiculate | Sponge | 39 and 40 |
| Crambescidin 359 (19) crambescidin acid (20) | 3D7 = 20.75 and 10.43 μM Dd2 = 18.67 and 5.03 μM |
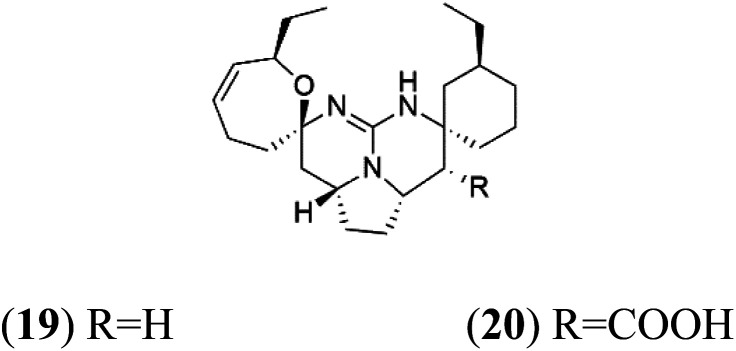
|
|||
| Fromiamycalin (21) | 3D7 = 0.24 μM |
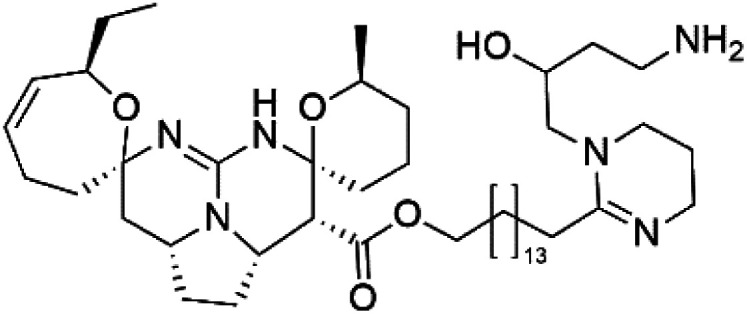
|
|||
| Unguiculin A (22) | 3D7 = 12.86 μM |
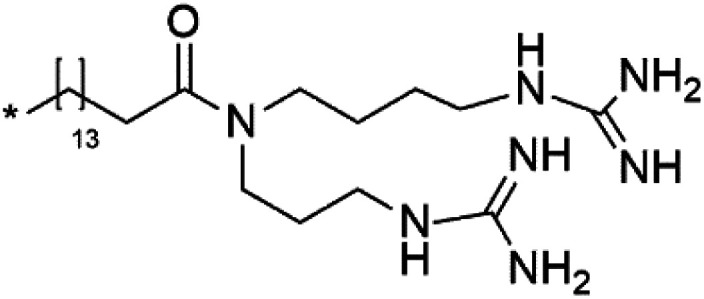
|
|||
| Ptilomycalins E (23) Ptilomycalins F (24) Ptilomycalins G (25) | 3D7 = 0.35, 0.23, and 0.46 μM |
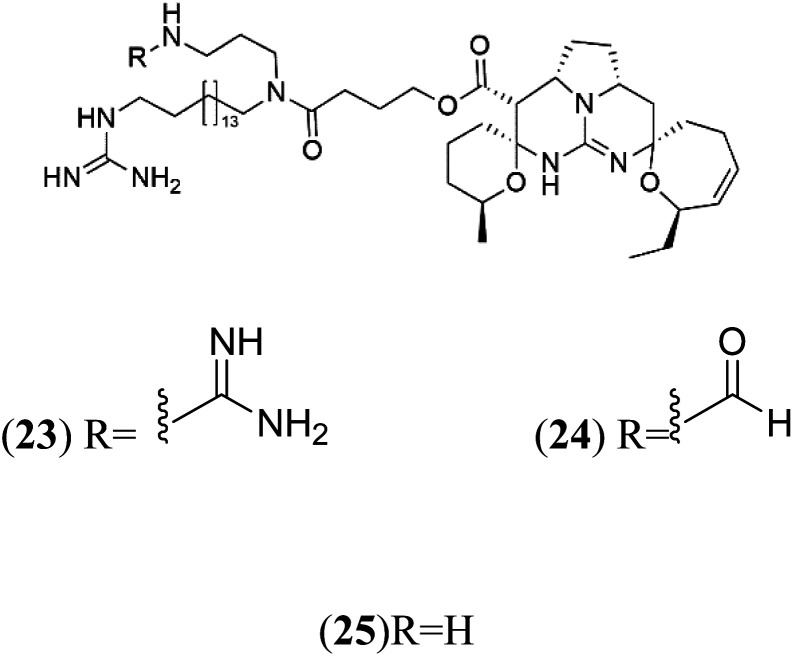
|
41 | ||
| Ptilomycalins H (26) | 3D7 = 0.46 μM |
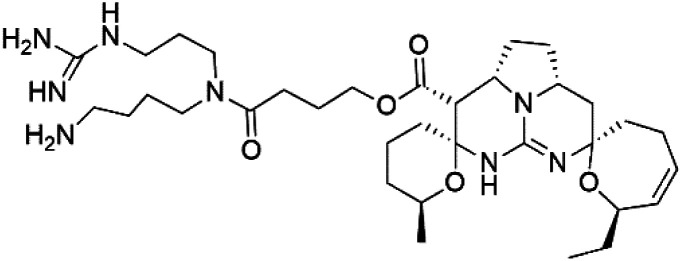
|
|||
| Opacaline B (27) Opacaline C (28) | K1 range of 2.5–14 μM |
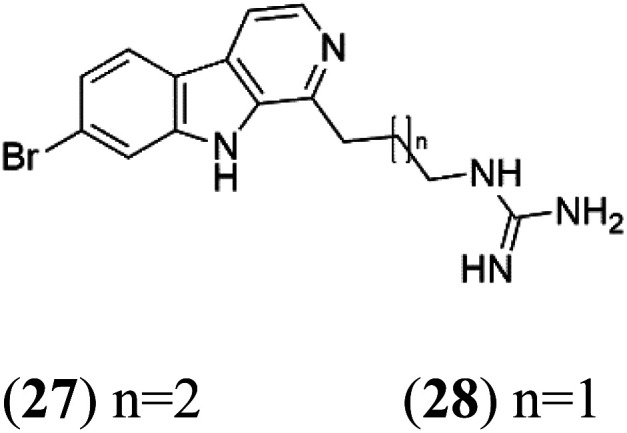
|
Pseudodistoma opacum | New Zealand ascidian | 42 |
| Didemnidine A (29) Didemnidine B (30) | K1 = 0.047 μM |
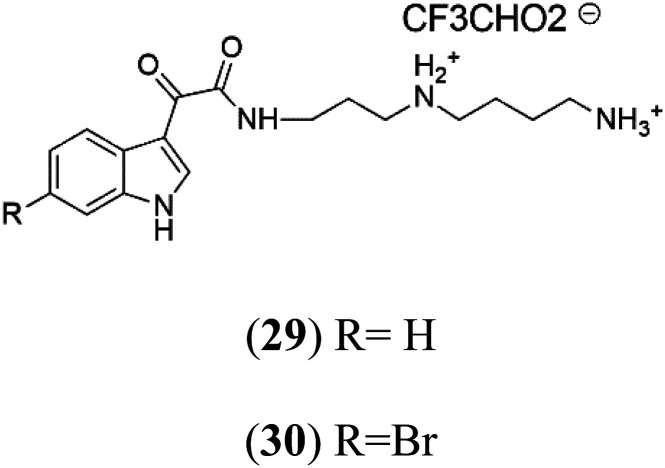
|
Ascidian Didemnum sp. | Marine tunicate | 43 |
| Salinosporamide A (31) | 3D7 = 11.4 nM FCB = 19.6 nM |
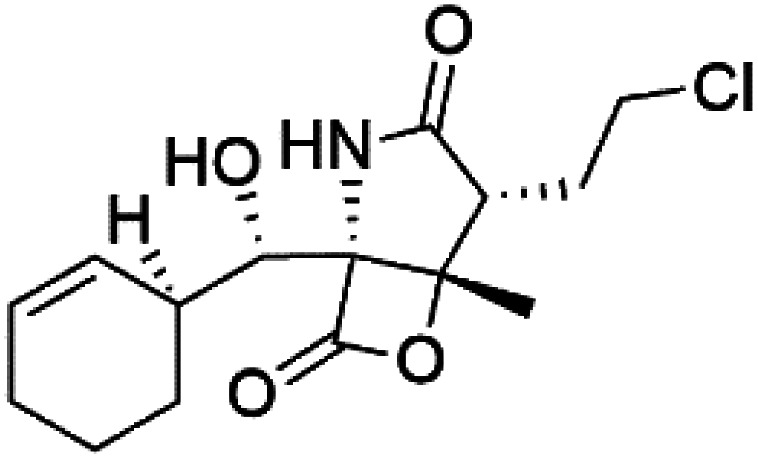
|
Salinispora tropica | Marine actinomycete bacteria | 44 |
| Psammaplysin H (32) Psammaplysin F (33) Psammaplysin G (34) | 3D7 = 0.41, 1.92, and 5.22 μM |
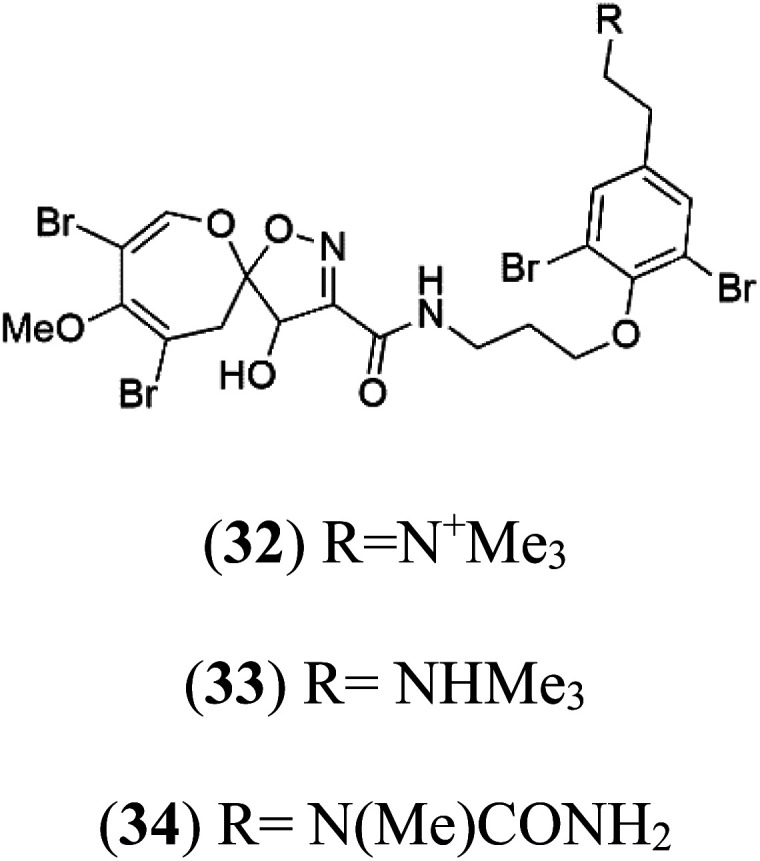
|
Aplysinella strongylata | Sponge | 52 |
| Ceratinadin E (35) | K1 = 0.9 μM |
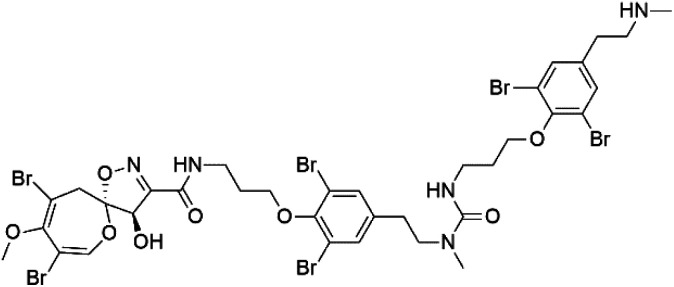
|
Okinawan Pseudoceratina | Sponge | 54 |
| Ceratinadin F (36) | K1 > 8.16 μM |
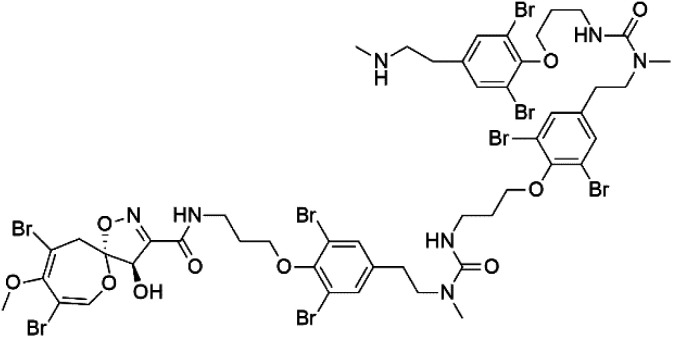
|
|||
| Tsitsikammamine C (37) | 3D7 = 13 nM Dd2 = 18 nM |
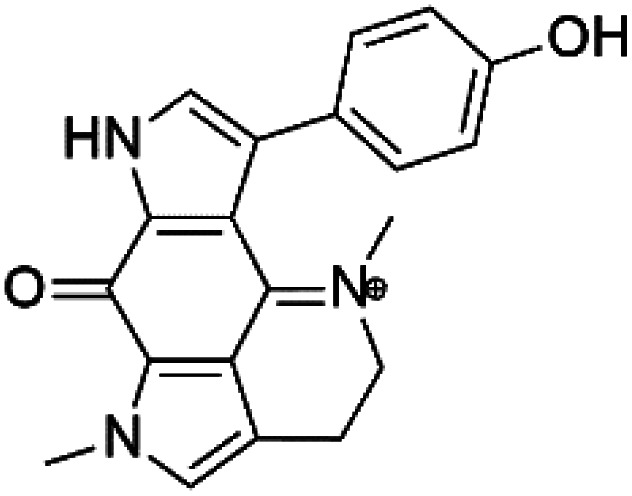
|
Zyzzya sp. | Sponge | 55 |
| Makaluvamine J (38) | 3D7 = 25 nM Dd2 = 22 nM |
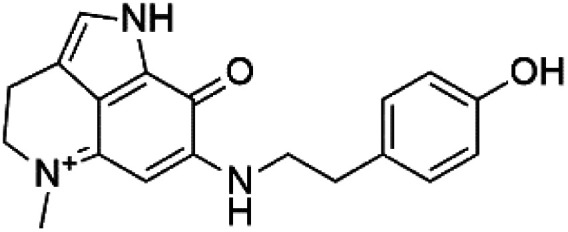
|
55 | ||
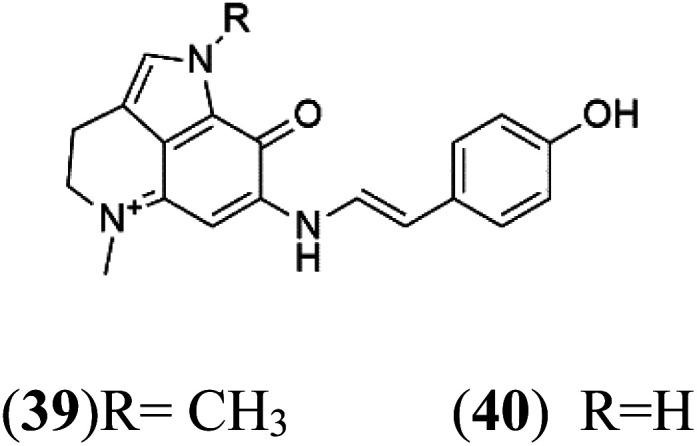
| Makaluvamine G (39) Makaluvamine L (40) | 3D7 = 36, 40 nM Dd2 = 39, 21 nM |
|
|||
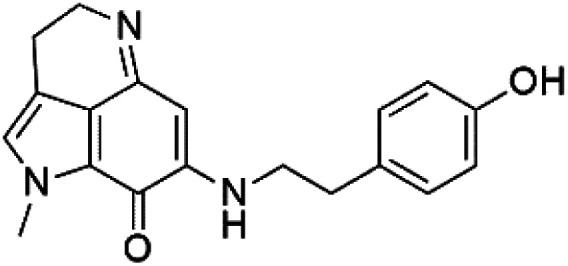
| Makaluvamine K (41) | 3D7 = 396 nM Dd2 = 300 nM |
|
|||
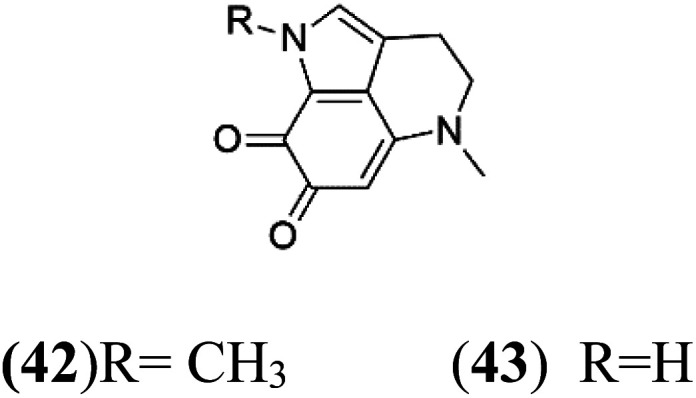
| Damirone A (42) Damirone B (43) | 3D7 = 1880 Dd2 = 360 nM |
|
|||
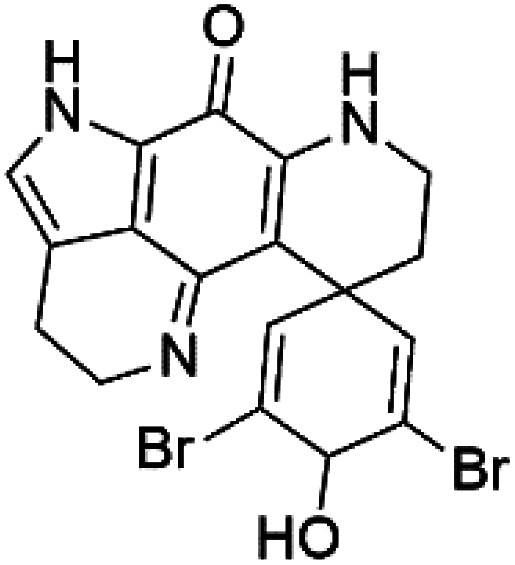
| Dihydrodiscorhabdin B (44) | D6 = 0.17 μM W2 = 0.13 μM |
|
Latrunculia sp. | Sponge | 56 |
| Discorhabdin Y (45) | EC50 = 0.5 μM |
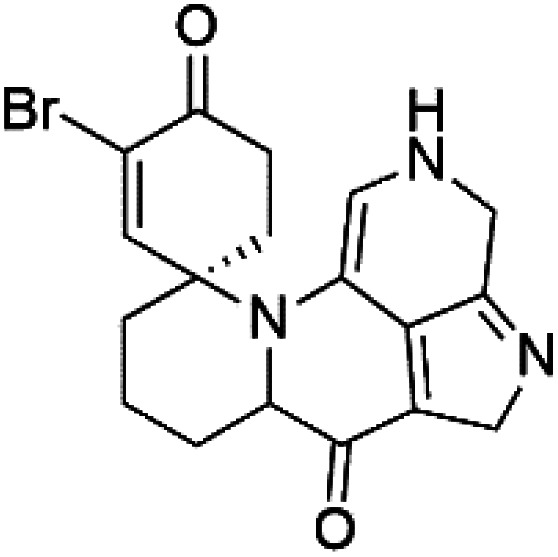
|
|||
| Discorhabdin A (46) | D6 = 0.05 μM W2 = 0.05 μM |
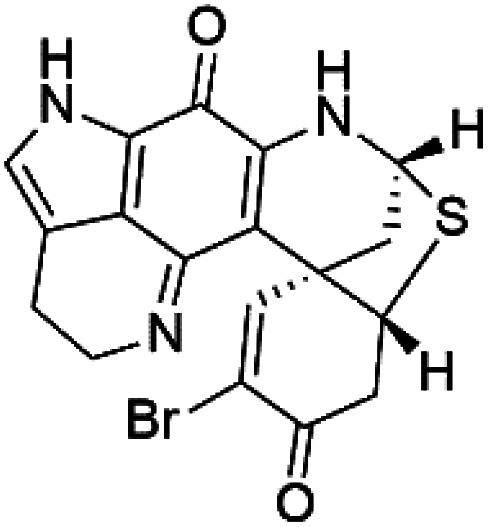
|
56 | ||
| Discorhabdin C (47) | D6 = 2.8 μM W2 = 2.0 μM |
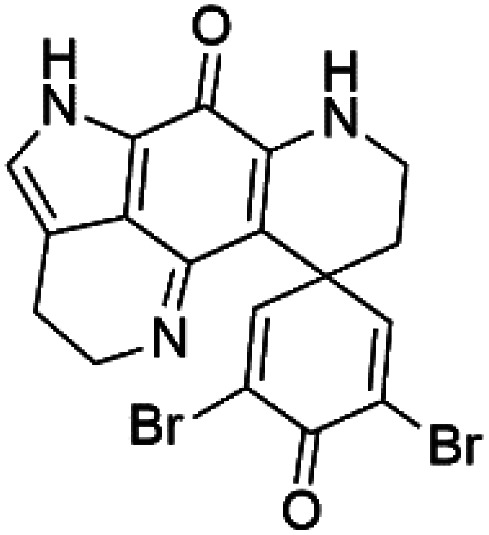
|
|||
| Discorhabdin E (48) | W2 = 0.2 μM |
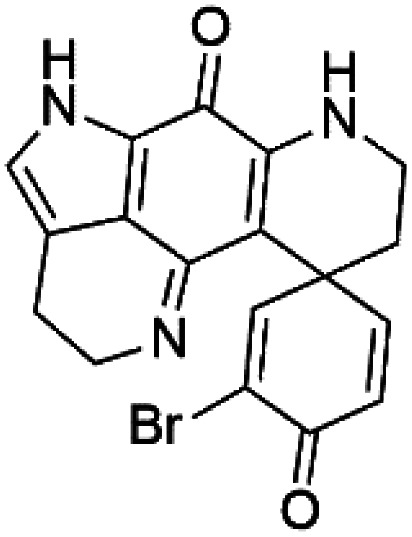
|
|||
| Discorhabdin L (49) | W2 = 0.13 μM |
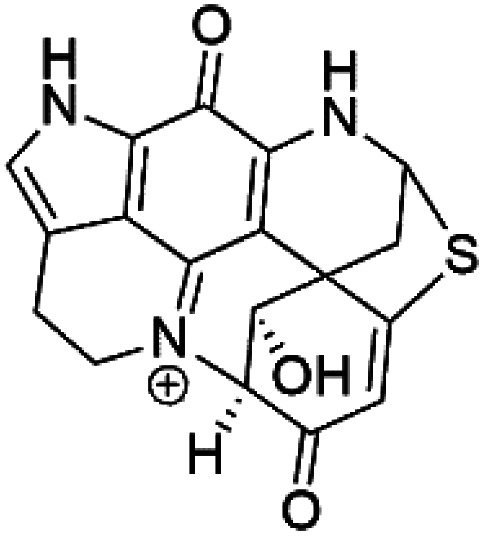
|
|||
| Dihydrodiscorhabdin C (50) | D6 = 0.17 μM W2 = 0.13 μM |
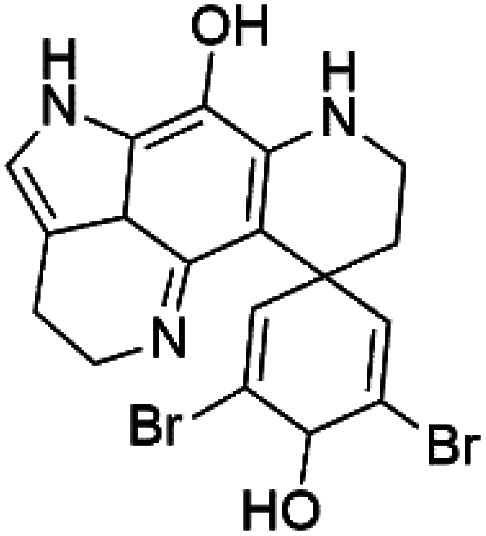
|
|||
| Girolline (51) | FCM29 = 0.13 μM |
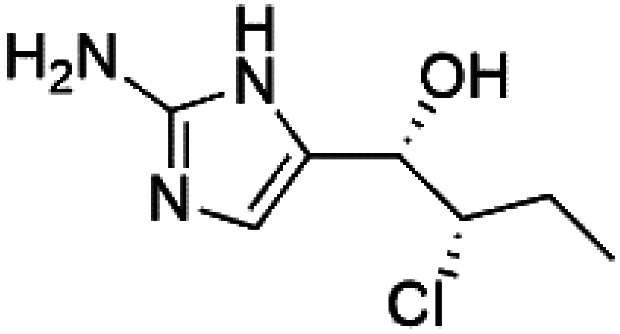
|
Cymbastela cantharella | Sponge | 58 |
| Thiaplakortone A (52) | 3D7 = 51 Dd2 = 6.6 nM |
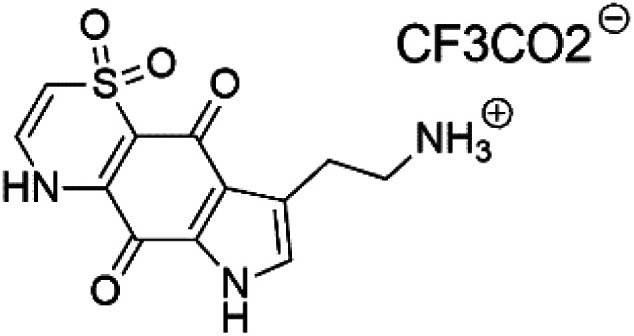
|
Plakortis lita | Sponge | 59 |
| Thiaplakortone B (53) | 3D7 = 650 Dd2 = 92 nM |
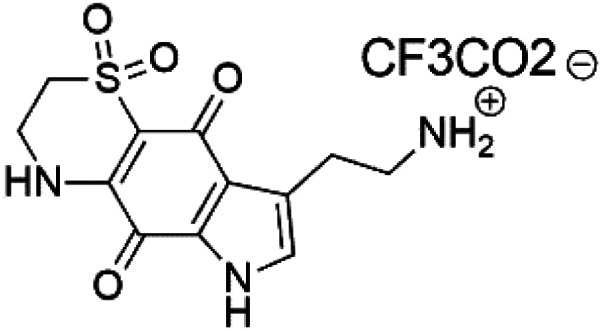
|
|||
| Thiaplakortone C (54) | 3D7 = 309 Dd2 = 171 nM |
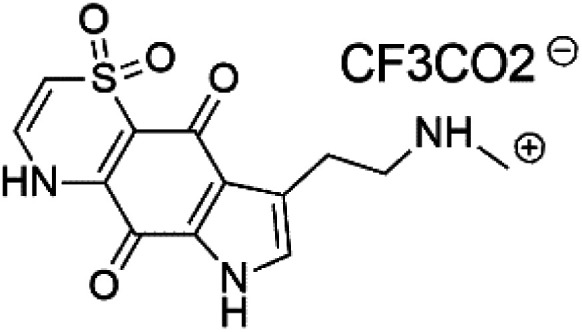
|
|||
| Thiaplakortone D (55) | 3D7 = 279 Dd2 = 159 nM |
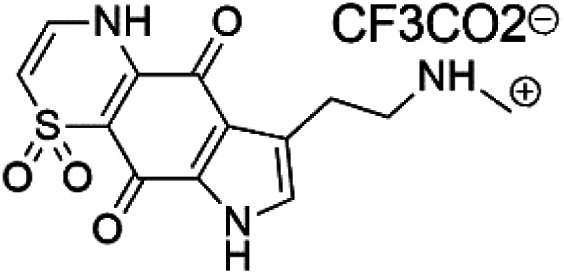
|
|||
| Monamphilectine A (56) | W2 = 600 nM |
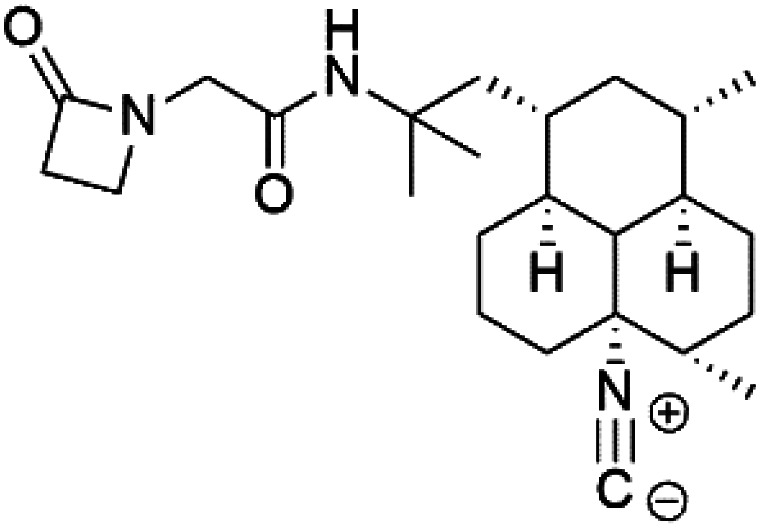
|
Hymeniacidon sp. | Sponge | 60 |
| Agelasine J (57) | FcB1 = 6.6 μM |
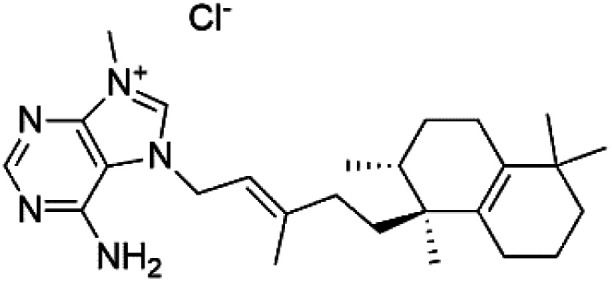
|
Agelas mauritiana | Sponge | 61 |
| Agelasine K (58) | FcB1 = 8.3 μM |
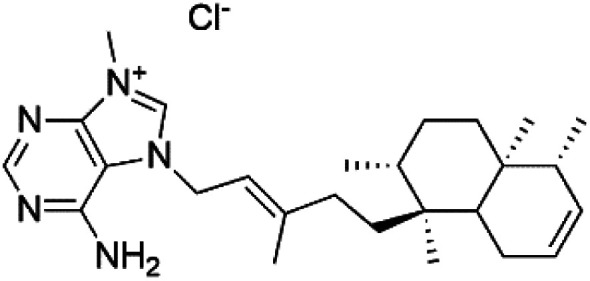
|
|||
| Agelasine L (59) | FcB1 = 18 μM |
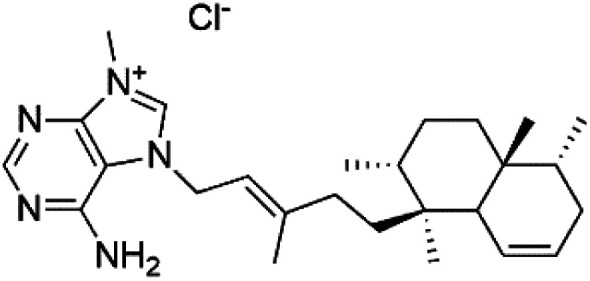
|
|||
| Netamine G (60) | NA |
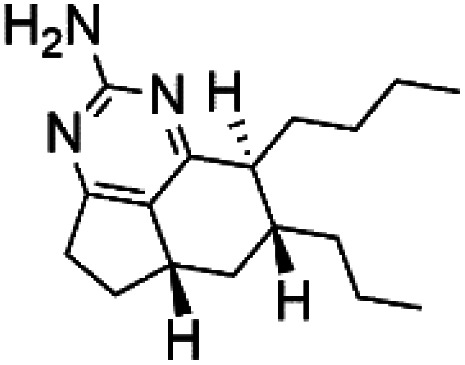
|
Biemna laboutei | Sponge | 62 |
| Netamine H (61) |
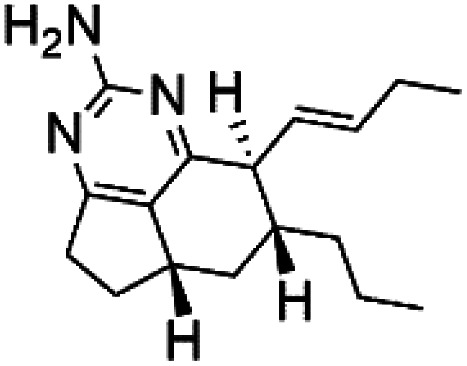
|
||||
| Netamine I (62) |
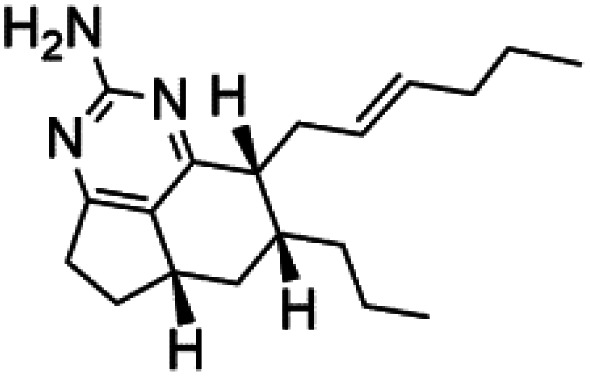
|
||||
| Netamine J (63) |
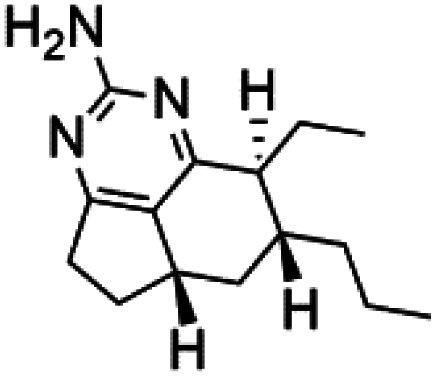
|
||||
| Netamine K (64) | (64) IC50 = 2.4 μM NA for the other compounds (65–67) |
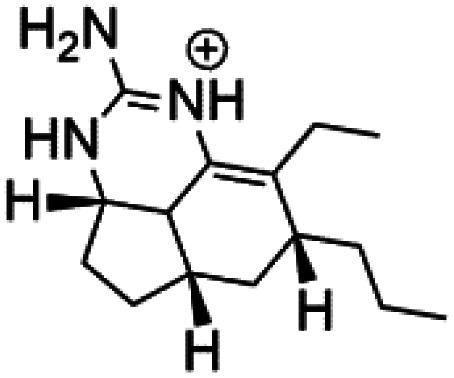
|
|||
| Netamine L (65) |
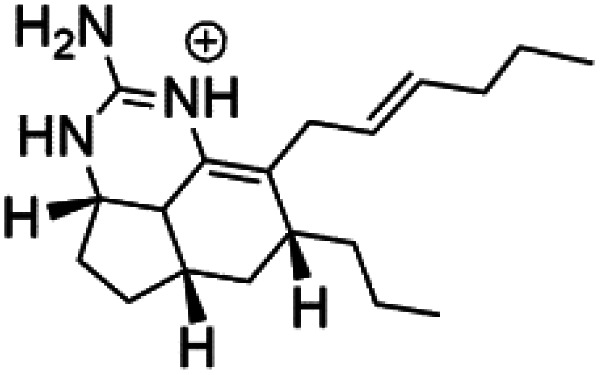
|
||||
| Netamine M (66) |
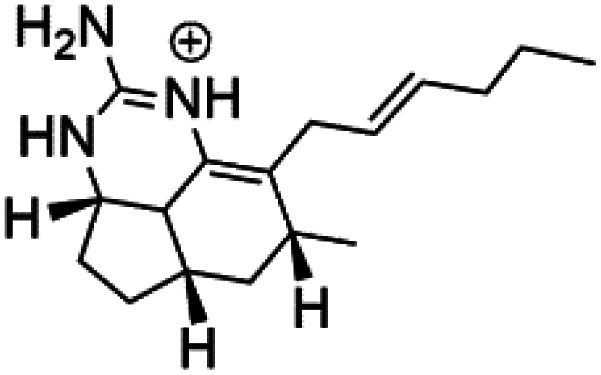
|
||||
| Netamine N (67) |
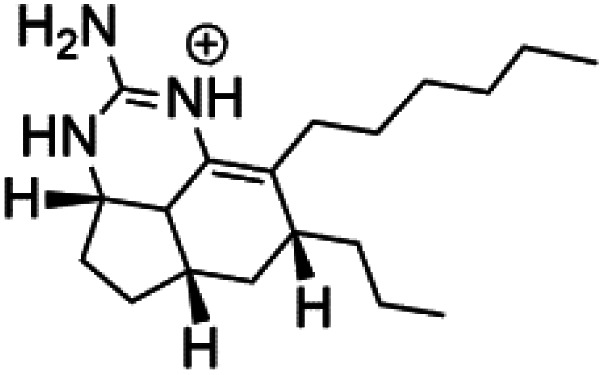
|
Biemna laboutei | Sponge |
NA:not available.
