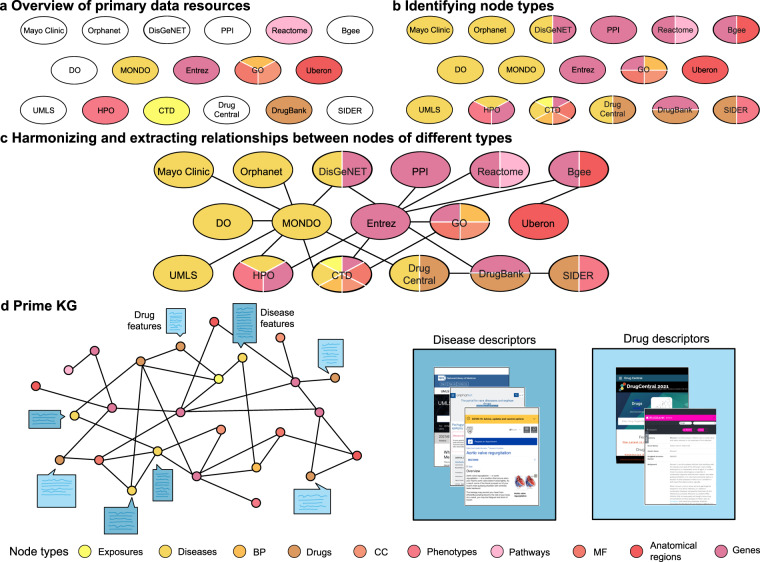
Fig. 2.
Building PrimeKG. The panels sequentially illustrate the process of developing the Precision Medicine Knowledge Graph. (a) Shown are 20 primary data resources curated to develop PrimeKG. The colors highlight which data records are used to uniquely identify each node type. For example, GO is colored by biological processes, cellular components, and molecular functions because GO terms are the unique identifiers used to define nodes for these three node types. (b) Primary resources are colored by each node type for which they possess information. For example, GO provides links from biological processes, cellular components, and molecular functions to genes. As a result, we add the fourth color to represent the gene/protein class. (c) Illustrated is the process of harmonizing these primary data records to extract relationships between node types. (d) The left side illustrates PrimeKG, and the right side shows all the textual sources of clinical information on drugs and diseases. The node type legend is consistent across the figure. Abbreviations - MF: molecular function, BP: biological process, CC: cellular component, PPI: protein-protein interactions, DO: disease ontology, MONDO: MONDO disease ontology, Entrez: Entrez gene, GO: gene ontology, UMLS: unified medical language system, HPO: human phenotype ontology, CTD: comparative toxicogenomics database, SIDER: side effect resource.