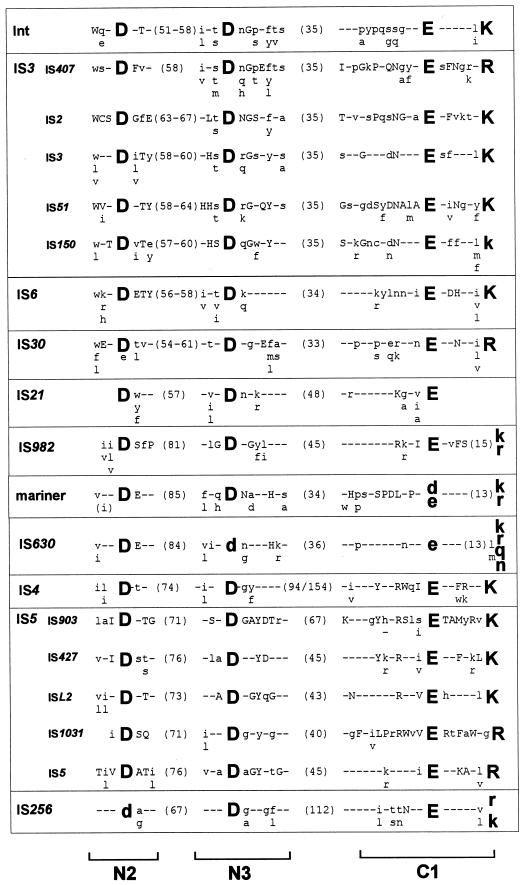
FIG. 3.
DDE consensus of different families. The alignments are derived from the groups presented in Table 1. Amino acids forming part of the conserved motif are shown as large bold letters. Capital letters indicate conservation within a family, and lowercase letters indicate that the particular amino acid is predominant. The numbers in parentheses show the distance in amino acids between the amino acids of the conserved motif. The retroviral integrase alignment is based on reference 266. The IS3 family is divided into the subgroups IS407, IS2, IS3, IS51, and IS150, as shown in Fig. 7B. The overall alignment (not shown) is essentially that obtained in reference 266. For IS21, see also reference 129; for mariner, see also references 87 and 295; for IS630, see also reference 87; for IS4 and IS5, see also reference 288. The IS5 family is divided into subgroups IS903, IS427, ISL3, IS1031, and IS5, as shown in Fig. 10. For IS256, see references 63 and 260. N2, N3, and C1 are regions defined in the IS4 transposon family (288).