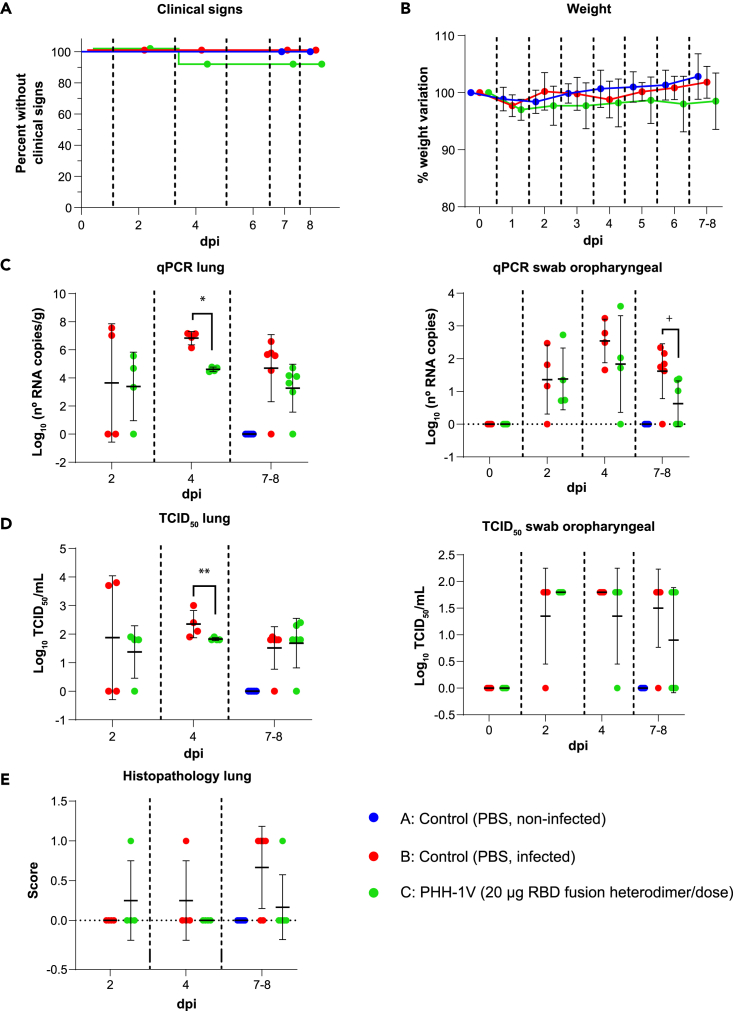
Figure 8.
Protective efficacy of PHH-1V vaccine in K18-hACE2 mice on challenge with SARS-CoV-2 Omicron BA.1 variant
(A) Group A (n = 8, 4F + 4M), group B (n = 18, 9F + 9M), and group C (n = 18, 9F + 9M), (A) Survival curves of animals from PHH-1V vaccinated groups and non-vaccinated groups. Survival analysis (Kaplan-Meier estimates and logrank test to compare groups) was performed to study differences in time to/before clinical signs and mortality.
(B) Mean weight change after Omicron BA.1 variant challenge calculated as a percentage of the pre-challenge weight in K18-hACE2 mice. A linear mixed effects model on the body weight change data was performed considering groups B and C. Points represent the average weight variation in each group and error bars depict a ± SD interval.
(C) SARS-CoV-2 RT-qPCR (number of copies) in the lungs and oropharyngeal swabs collected from challenged animals.
(D) Viral titers were determined using a standard TCID50 assay on positive samples of RT-qPCR. Negative samples are represented as 0 TCID50/mL. The detection limit was set at 1.8 TCID50/mL.
(E) Histopathological analyses from the lungs were determined for all animals. For each tissue sample, lesions were classified as previously assays. Lesions were evaluated with the following score: 0 (no lesion); 1 (mild lesion); 2 (moderate lesion); and 3 (severe lesion). All the samples correspond to 2 (D37), 4 (D39) and 7 dpi (D42 for males) or 8 days post infection (D43 for females); or at the time of euthanasia in animals reaching endpoint criteria before the scheduled euthanasia day. GLS models or Mann-Whitney tests were employed for the analysis of the RT-qPCR, TCID50 and histopathological data depending on verification of assumptions. Each data point represents an individual mouse value, with bars representing the mean ± SD. Statistically significant differences between groups are indicated with a line on top of each group: ∗p<0.05; ∗∗p<0.01; +0.05<p<0.1. DPI: days post infection. See also Figure S2.