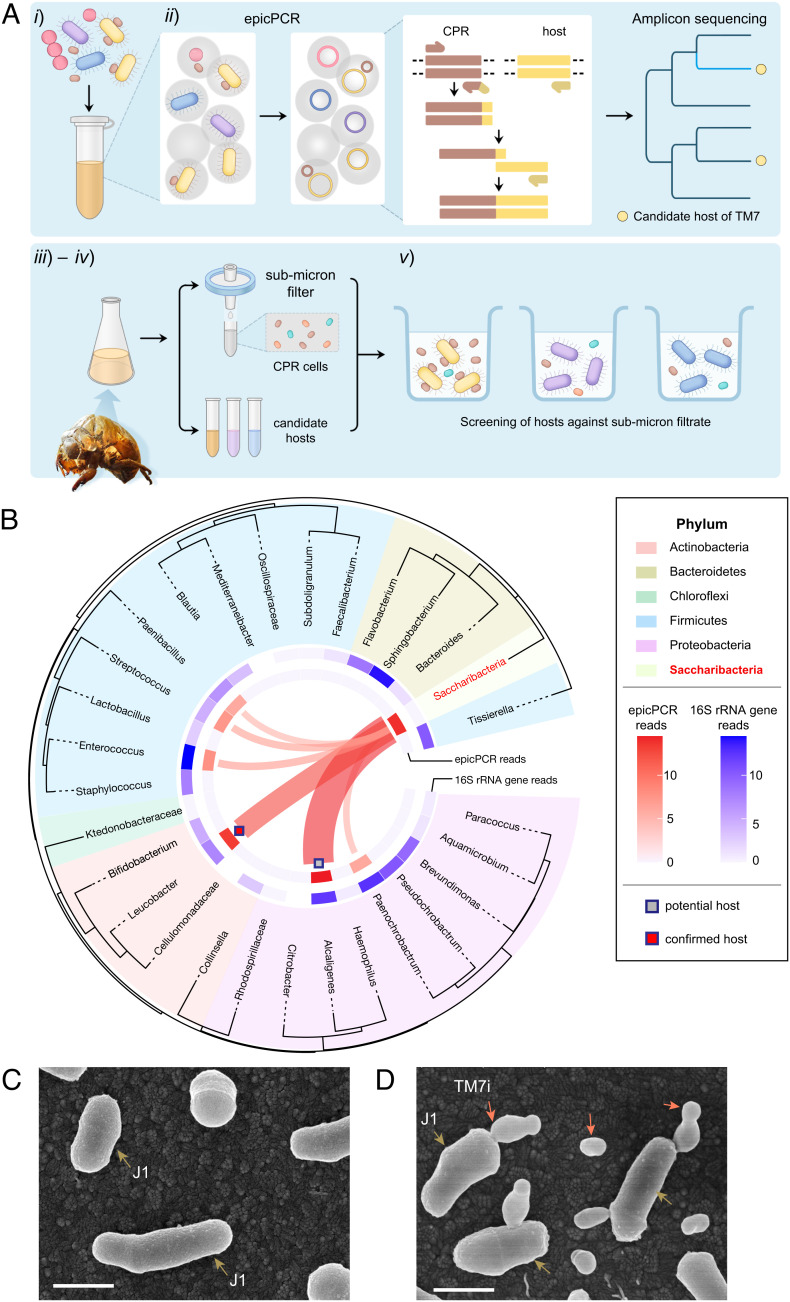
Fig. 1.
Gene-targeted episymbiont cultivation leads to the isolation of the first insect-associated Saccharibacteria (TM7) phylotype. (A) The workflow of gene-targeted TM7 symbiont isolation include five steps: (i) prepare microbial suspension, (ii) profile the TM7‒host association using epicPCR (emulsion, paired isolation, and concatenation PCR), (iii) selectively cultivate the candidate hosts, (iv) filtrate reinfection, and (v) identification and recovery of TM7‒host symbionts. (B) EpicPCR-informed TM7‒host associations in the enrichment community of Cicadae Periostracum. We built the phylogenetic tree shown in the outer circle with 16S rRNA gene sequences. Taxon groups are depicted as colored segments. The relative abundance of 16S rRNA amplicon sequencing and epicPCR-informed associations are annotated in the inner blue and red rings, respectively. Ribbons connecting to TM7 indicate potential TM7‒host associations. (C and D) The information obtained through epicPCR led to the cultivation of Saccharibacteria TM7i with the microbial host L. aridicollis J1, as shown in the scanning electron microscopy (SEM) images. (Scale bar, 500 nm.) (C) Pure culture of actinobacterial host J1. Brown arrows indicate J1 cells. (D) TM7i‒J1 symbiont. Red arrows indicate either planktonic or host-adhered TM7i cells.