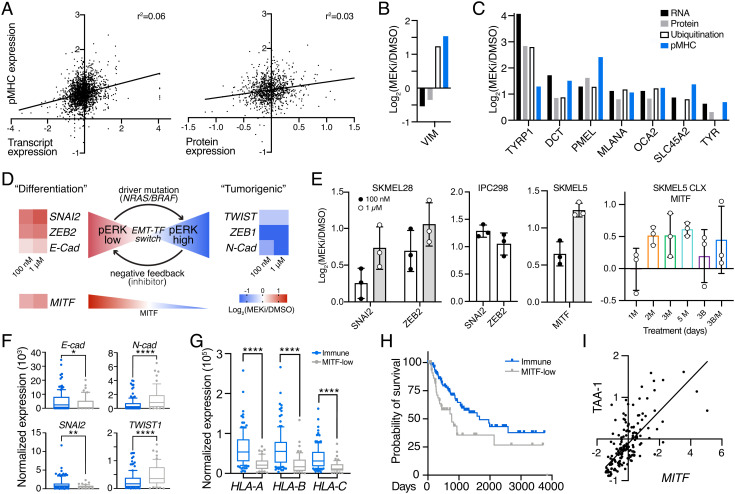
Fig. 3.
MEKi-induced EMT phenotype switching enhances TAA expression. (A) Correlation between pMHC expression and transcript expression (Left)/protein expression (Right) in SKMEL5 cells +/− 100 nM MEKi. Values represent Log2(MEKi/DMSO). (B), (C) Changes in RNA, protein, ubiquitination, and pMHC expression of selected targets. (D) Schematic of EMT-TF switching, with changes in transcription (SKMEL5 +/− 100 nM or 1 uM binimetinib) shown next to gene names. (E) Maximum average change in expression of pMHCs derived from EMT-related source proteins (n = 3). M = MEKi, B = BRAFi. Error bars = SD. (F–G) Normalized expression of select genes in NRAS/BRAF mutant immune (blue, n = 122) and MITF-low (gray, n = 54) classified tumors. *P < 0.05, **P < 0.01, ****P < 0.0001, unpaired 2-tailed t test. (H) Kaplan Meier curve of BRAF/NRAS immune and MITF-low melanomas. P = 0.012, log-rank test. (I) Correlation in expression of MITF and average TAA-1 gene set Z-scored expression for immune/MITF-low tumors, r = 0.723, P < 0.0001 (two-tail).