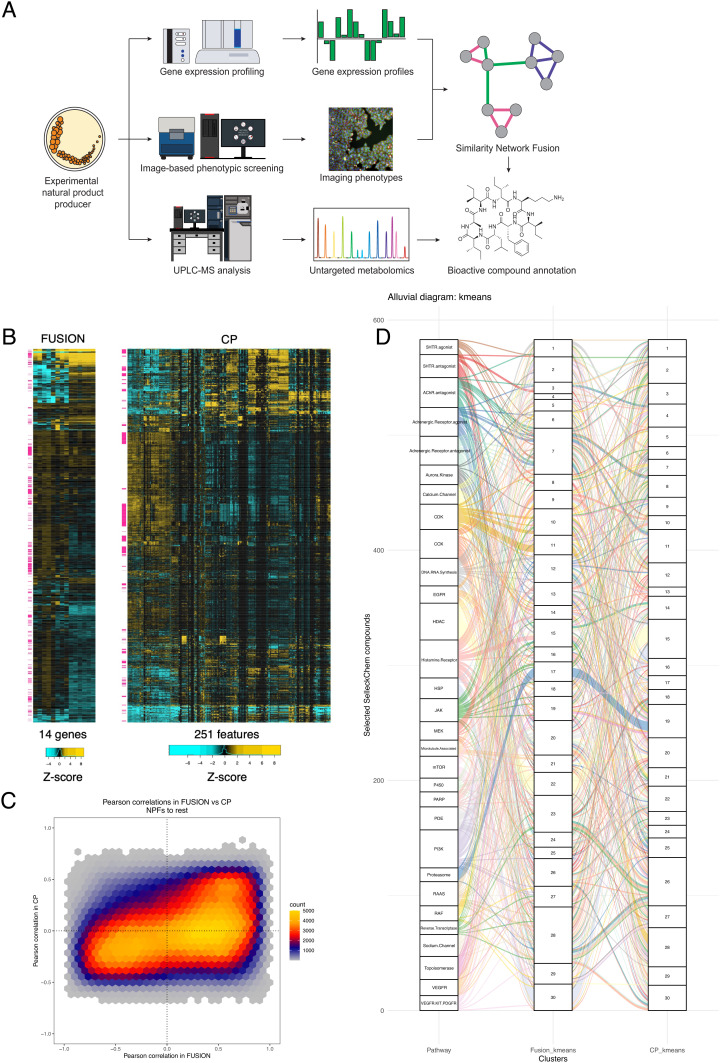
Fig. 1.
Overview of screening platforms and initial data collection. (A) Experimental outline. Natural product fractions are isolated from marine bacteria, screened through two biological screening platforms (FUSION and CP), and subjected to high-resolution mass spectrometry-based metabolomics profiling. FUSION and CP data are integrated using SNF, which then provides biological annotation on individual metabolites identified. (B) Two- way hierarchical clustering of Z-scores from FUSION and CP using Euclidean distance and complete linkage. NPFs are indicated with pink flags. (C) Heat-scatter hexplot comparing Pearson correlations between NPFs and all other perturbagens in FUSION vs. CP. (D) Alluvial diagram comparing k-means clustering of chemicals in the top 30 largest target classes in FUSION and CP. Each target class is represented by a different color.