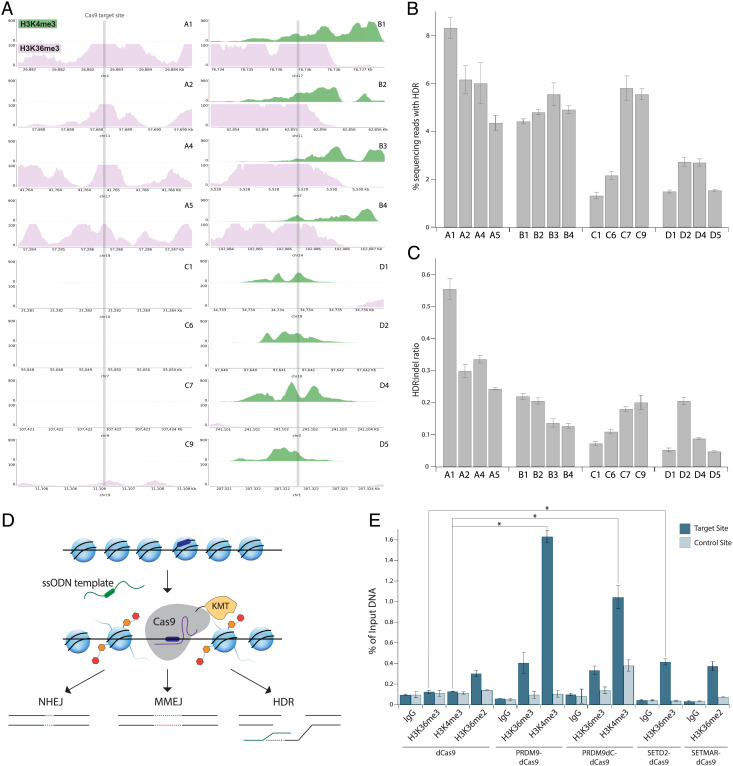
Fig. 1.
Endogenous histone modifications mediate DNA repair pathway choice. (A) University of California, Santa Cruz Genome Browser tracks showing normalized endogenous levels of H3K4me3 (green) and H3K36me3 (pink) at target site ± 1.5 kb based on ENCODE ChIP-seq datasets from HEK293 cells. Each plot spans 3 kb, and the Y-axis reports the negative log P value for peak enrichment. (B) HDR frequency measured by NGS at genomic sites with varying endogenous H3K4me3 and H3K36me3 enrichment in HEK293T cells transfected with plasmids expressing Cas9 and sgRNA along with an ssODN template with 50-nt homology arms. Data represent mean ± SD (n = 3). (C) HDR:indel ratio measured by NGS at genomic sites with varying endogenous H3K4me3 and H3K36me3 enrichment in HEK293T cells transfected with plasmids expressing Cas9 and sgRNA along with an ssODN template with 50-nt homology arms. Data represent mean ± SD (n = 3). (D) Schematic of Cas9-histone lysine methyltransferase (KMT) fusion protein activity. Cas9 fusion protein is guided by an sgRNA (purple) to the DNA target site (dark blue), which may be embedded within a heterochromatic region. Cas9 fusion protein deposits histone marks (orange and red) at the target site, which influence the choice of DNA repair pathway following Cas9-induced DSBs. (E) H3K36me3, H3K4me3, and H3K36me2 enrichment shown as a percentage of input DNA measured by ChIP-qPCR at site C7 in HEK293T cells 3 dpt. Cells were transfected with plasmids expressing the dCas9 fusion proteins and sgRNA. Data represent mean ± SD (n = 2). *P < 0.05, determined by Student’s two-tailed t test.