Figure 1.
An integrated multi-omic resource in 15 diploid human cell types
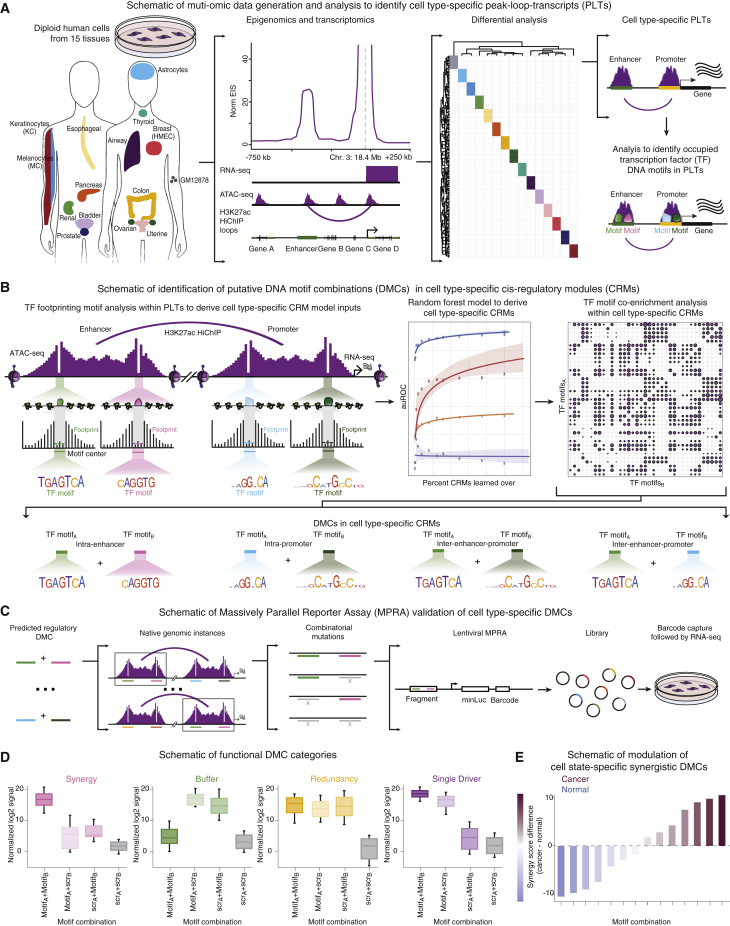
(A) Workflow for cell-type-specific ATAC peaks, HiChIP loops, and target gene transcripts (PLTs) across 15 diploid human cell types.
(B) Schematic of transcription factor (TF) footprinting analysis within PLTs to identify inputs for a random-forest model to derive cell type CRMs. Co-enrichment analysis within CRMs extracted DMCs.
(C) Native genomic instances of putative intra-enhancer and intra-promoter DMCs were tested via MPRA. Combinatorial mutations were used to assess cooperativity of DMCs in a lentiviral setup.
(D) Schematic of MPRA-validated functional categories of DMC interactions.
(E) Schematic bar plot comparing synergistic DMC MPRA activity of normal and cancer-derived DMCs in corresponding cell types.